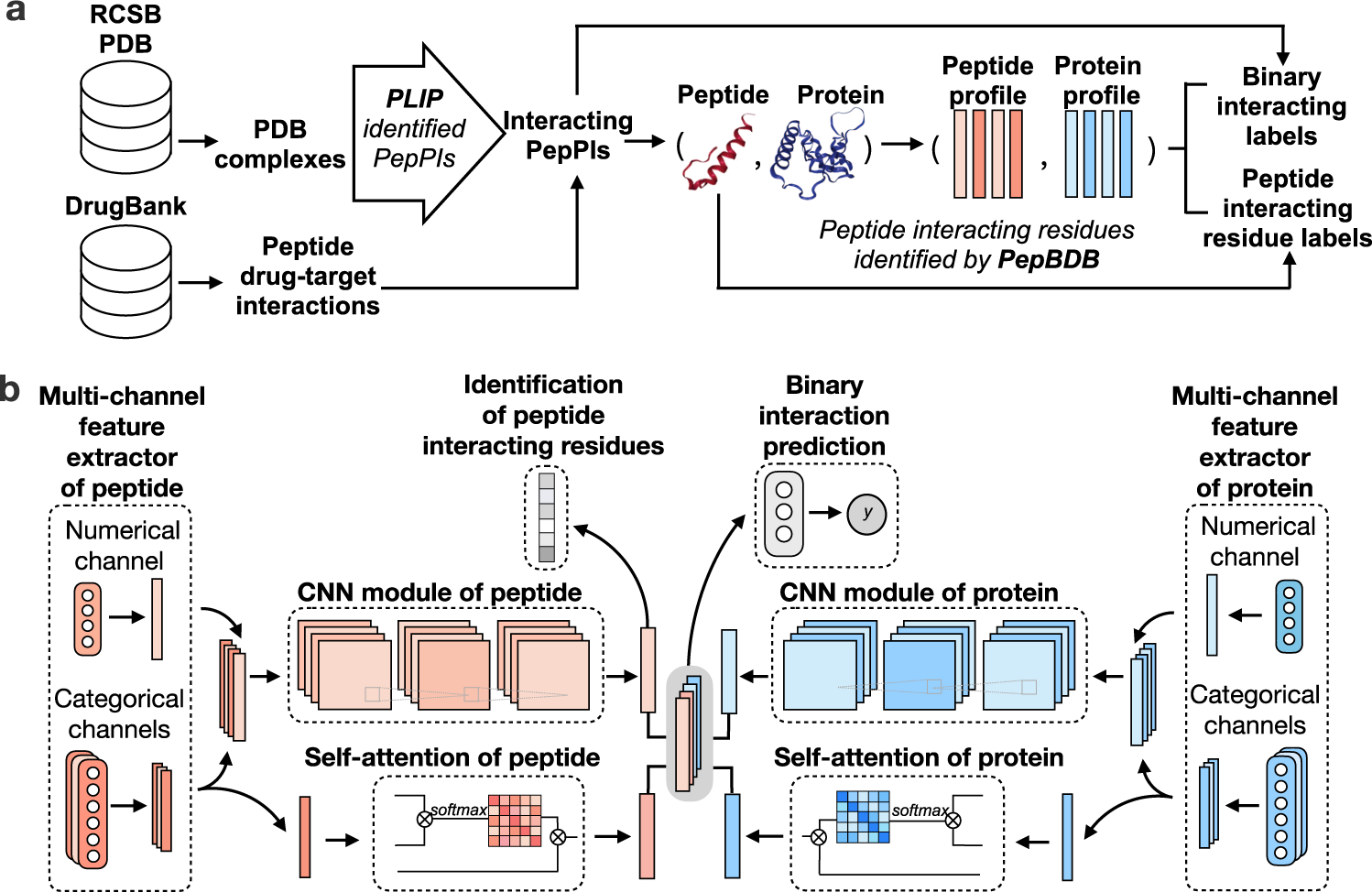
A deep-learning framework for multi-level peptide–protein interaction prediction | Nature Communications
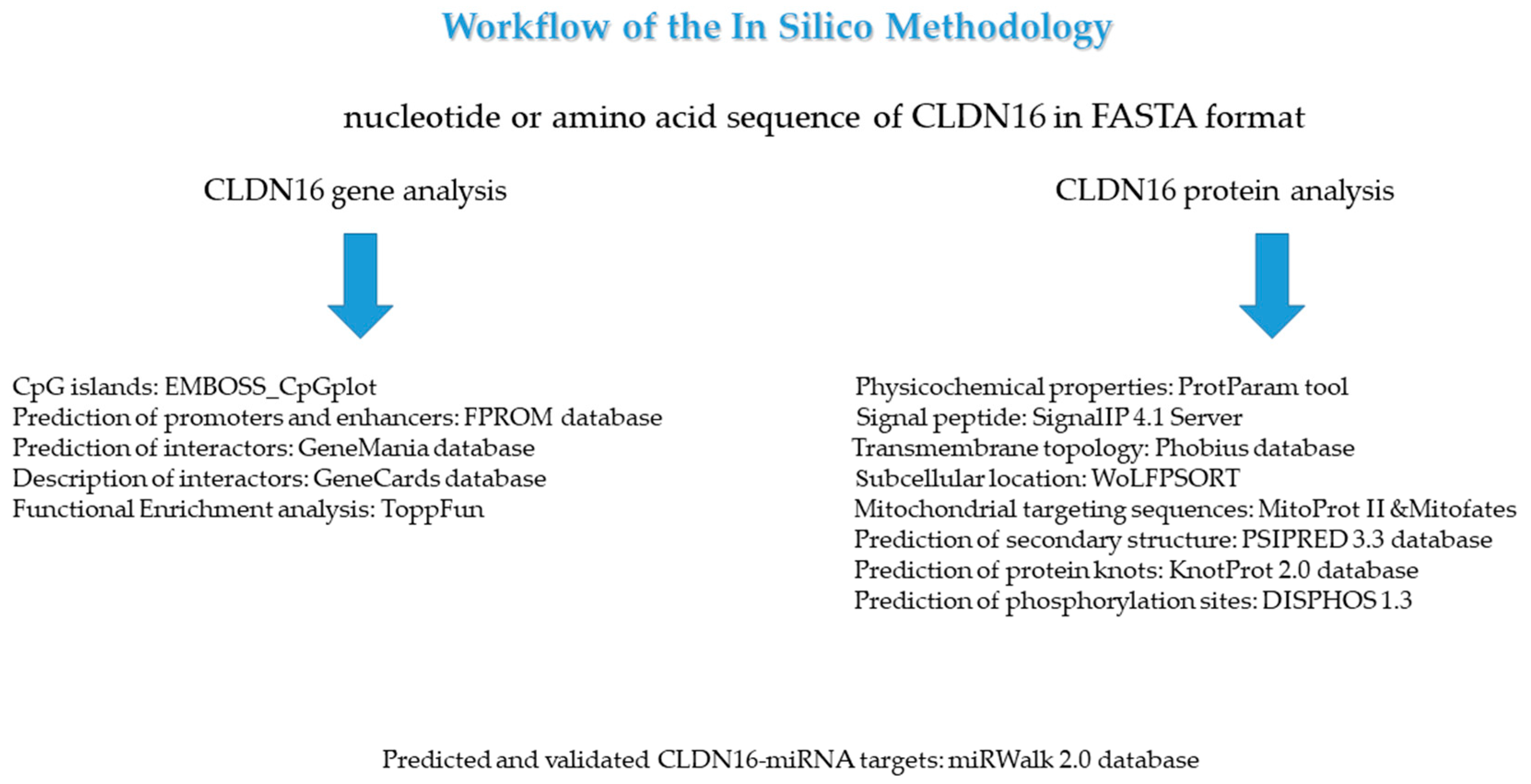
Medicina | Free Full-Text | In-Depth Bioinformatic Study of the CLDN16 Gene and Protein: Prediction of Subcellular Localization to Mitochondria

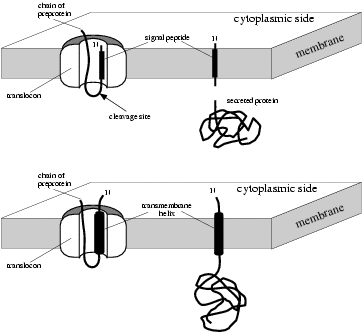
Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide | eLife

Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage - ScienceDirect
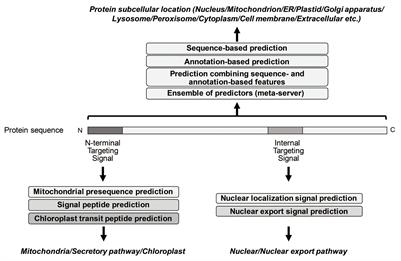
Frontiers | Tools for the Recognition of Sorting Signals and the Prediction of Subcellular Localization of Proteins From Their Amino Acid Sequences

Transmembrane Helix Prediction always reports no transmembrane regions found - QIAGEN Digital Insights
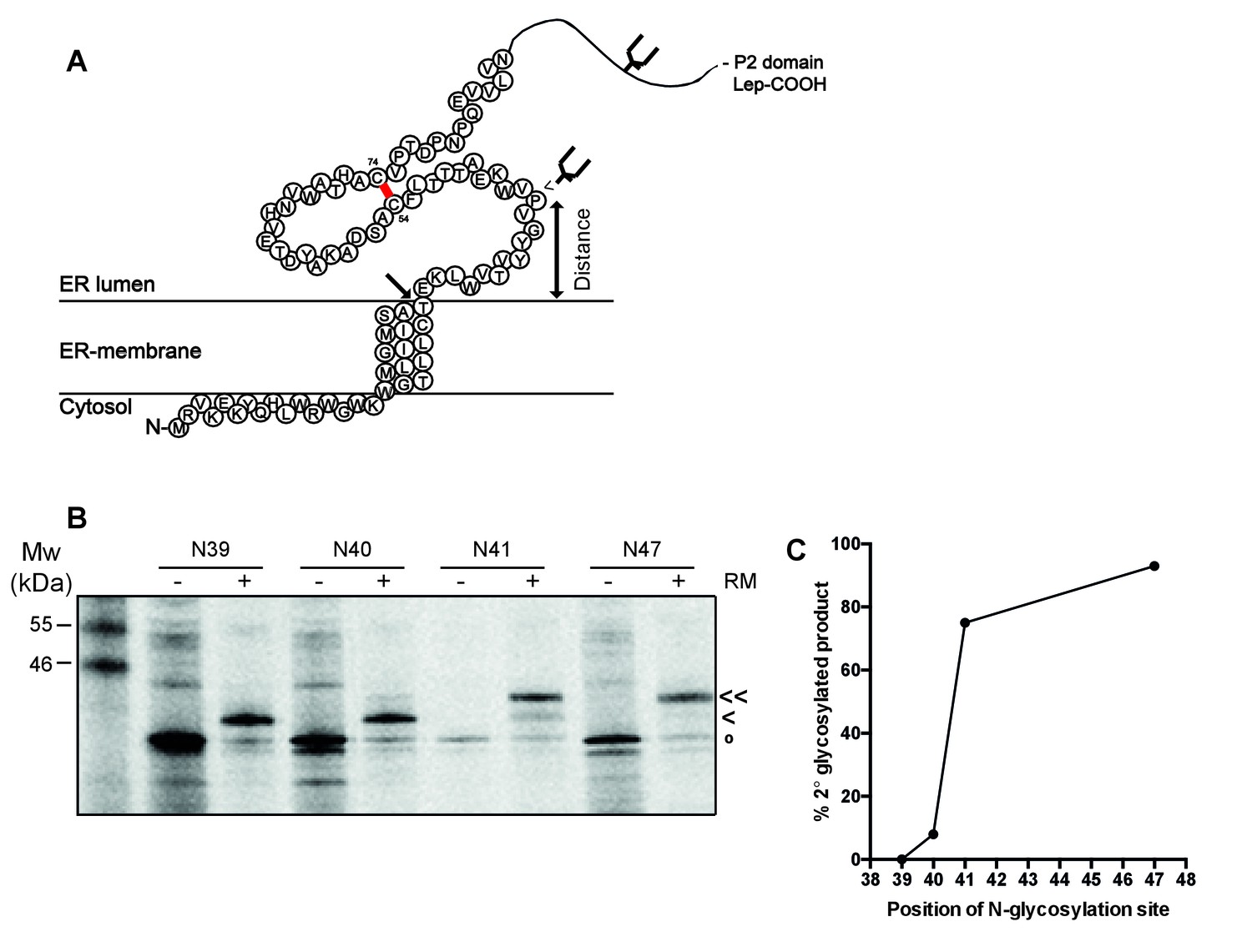
Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide | eLife

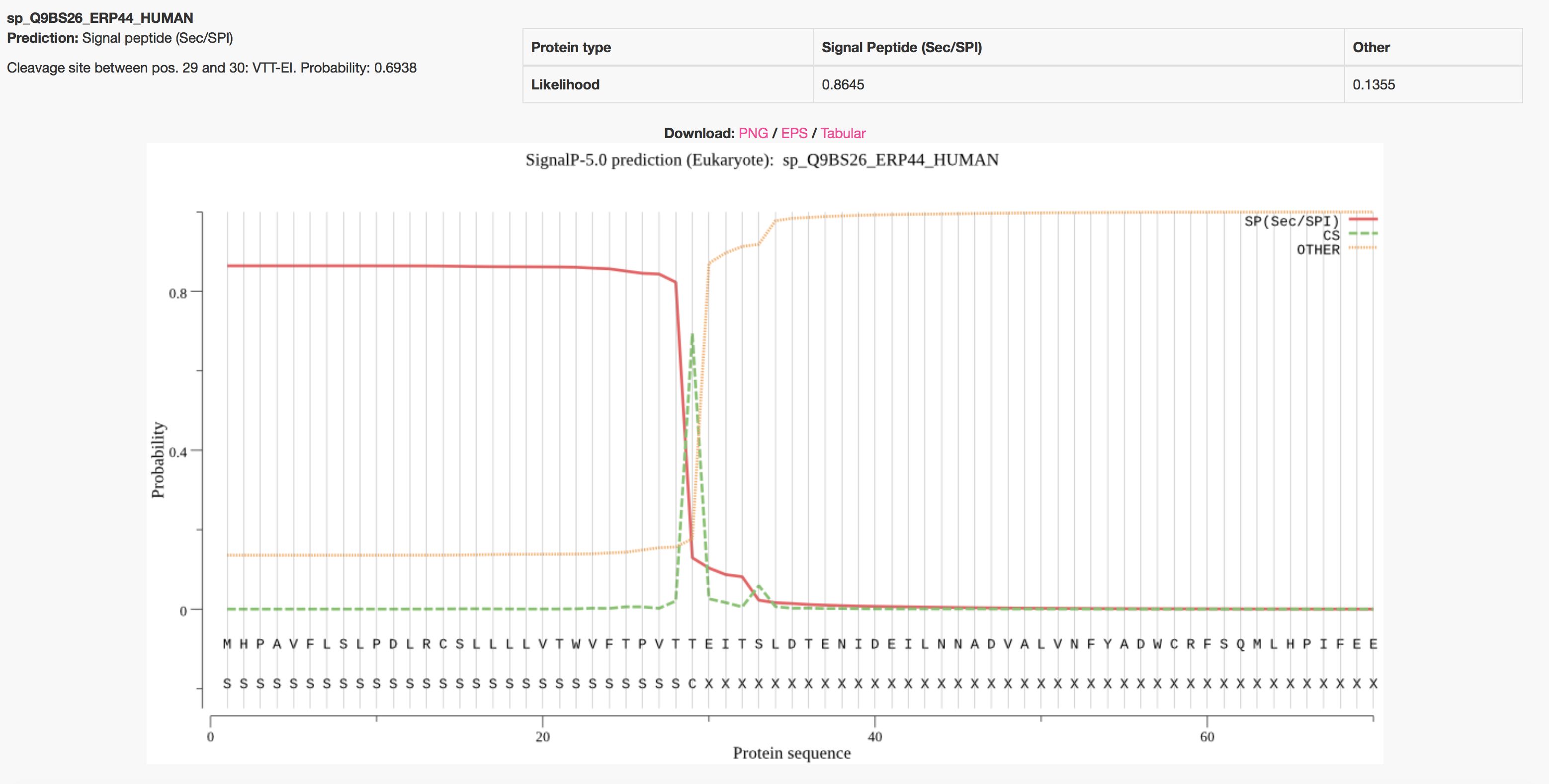
Signal peptide prediction. Signal peptide sequence was predicted using... | Download Scientific Diagram

Proteome-Scale Screening to Identify High-Expression Signal Peptides with Minimal N-Terminus Biases via Yeast Display | ACS Synthetic Biology
![PDF] Nuclear export signals (NESs) in Arabidopsis thaliana : development and experimental validation of a prediction tool | Semantic Scholar PDF] Nuclear export signals (NESs) in Arabidopsis thaliana : development and experimental validation of a prediction tool | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/b5e2c3d4eec70842f8f091b442a39f777b1ea9be/22-Figure2.1-1.png)

![PDF] PrediSi: prediction of signal peptides and their cleavage positions | Semantic Scholar PDF] PrediSi: prediction of signal peptides and their cleavage positions | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/296e275b6aae81a508d925835d6617c499fbe0ec/4-Figure2-1.png)