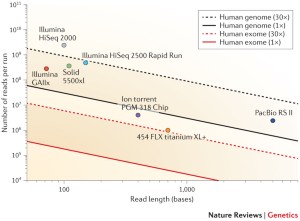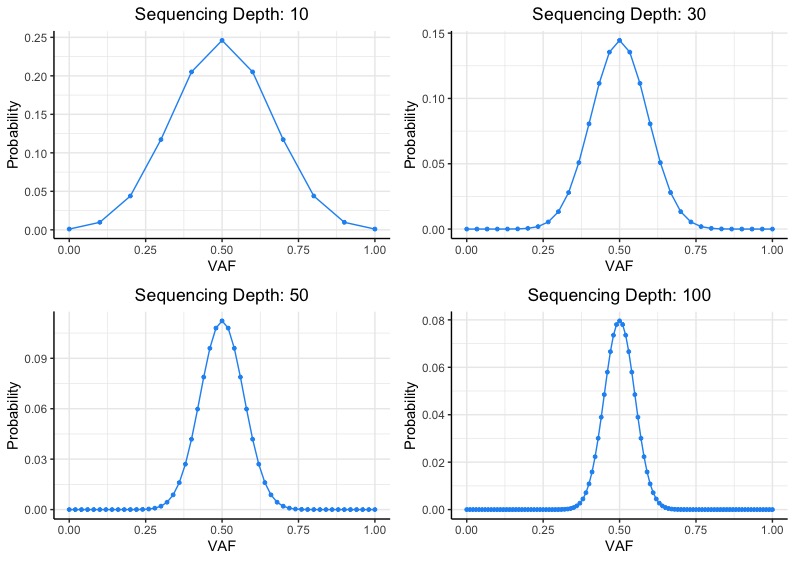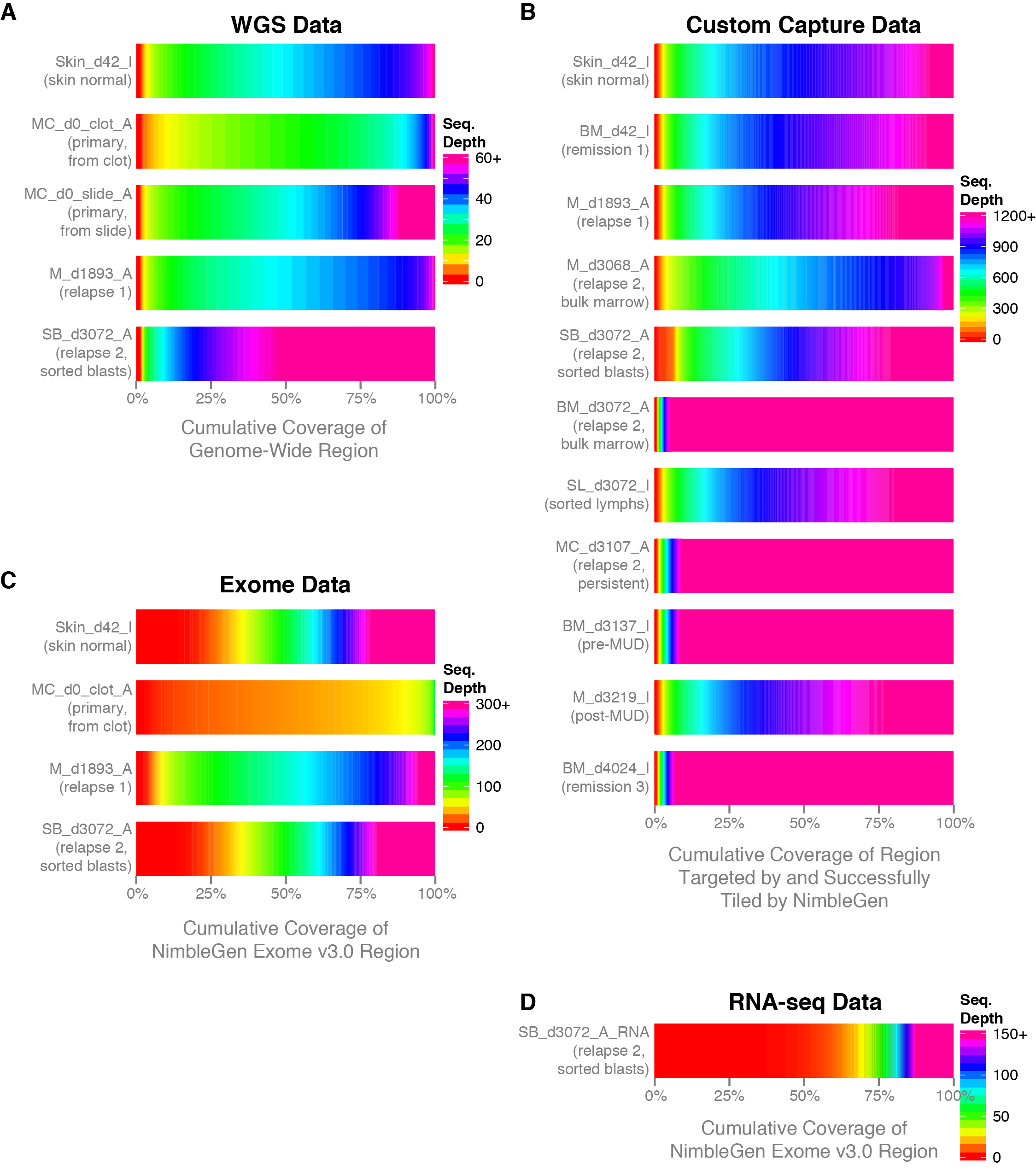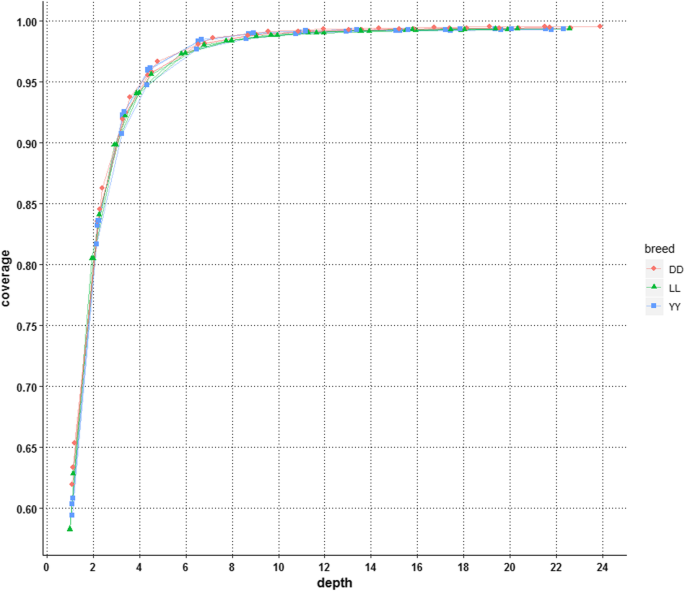
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text

Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers

Recommendations for accurate genotyping of SARS-CoV-2 using amplicon-based sequencing of clinical samples - ScienceDirect

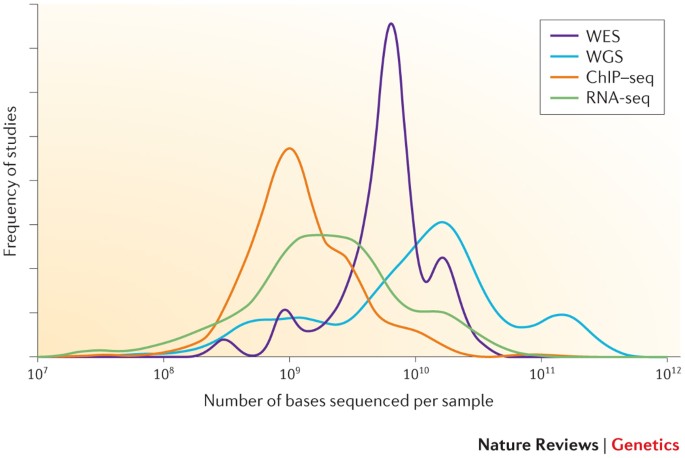
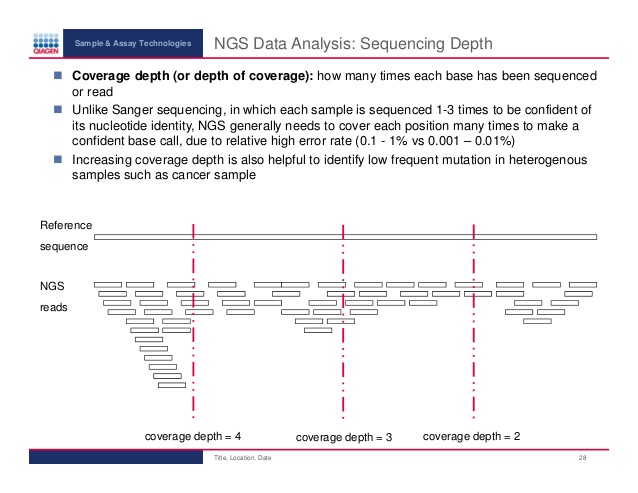
A beginner's guide to low‐coverage whole genome sequencing for population genomics - Lou - 2021 - Molecular Ecology - Wiley Online Library
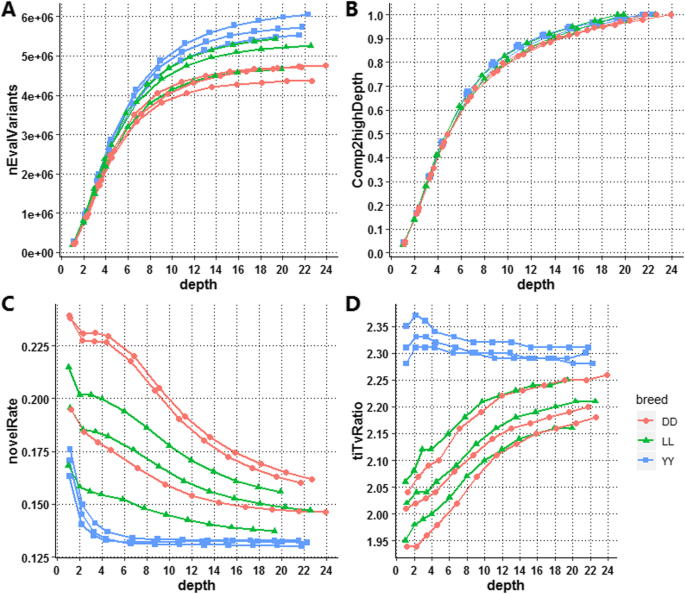
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text