Influences on gene expression in vivo by a Shine–Dalgarno sequence - Jin - 2006 - Molecular Microbiology - Wiley Online Library

Extending the Spacing between the Shine–Dalgarno Sequence and P-Site Codon Reduces the Rate of mRNA Translocation - ScienceDirect
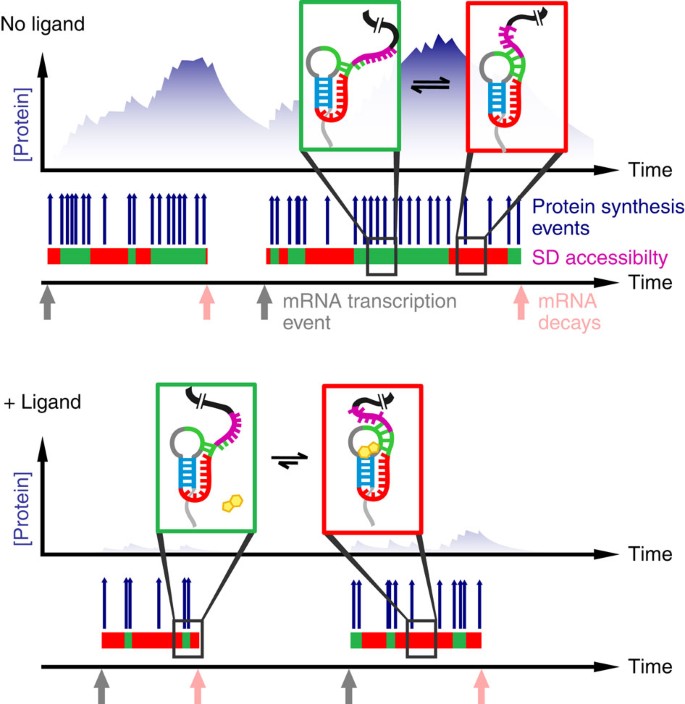
The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts | Nature Communications
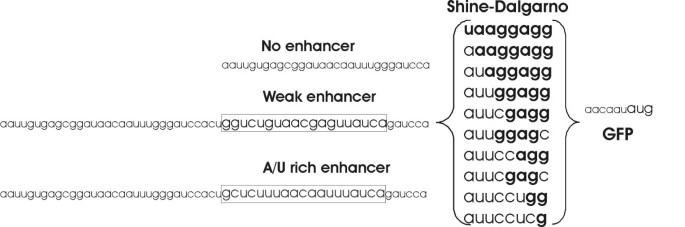
Translation initiation region sequence preferences in Escherichia coli | BMC Molecular Biology | Full Text

Sequence entropy quantifies genome-wide SD sequence utilization. (A),... | Download Scientific Diagram

The possible dual impacts of Shine-Dalgarno (SD) sequences on protein... | Download Scientific Diagram

The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts | Nature Communications

SD sequence usage is variably defined in the literature and differs... | Download Scientific Diagram

A model of SD sequence and aSD interactions. (A) The free 39 end of SSU... | Download Scientific Diagram
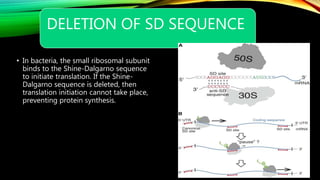
![PDF] Shine-Dalgarno Sequences Play an Essential Role in the Translation of Plastid mRNAs in Tobacco | Semantic Scholar PDF] Shine-Dalgarno Sequences Play an Essential Role in the Translation of Plastid mRNAs in Tobacco | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/994e2f8be82562f98a5c4bc645df7d19baf22bfb/2-Figure1-1.png)
PDF] Shine-Dalgarno Sequences Play an Essential Role in the Translation of Plastid mRNAs in Tobacco | Semantic Scholar

Influences on gene expression in vivo by a Shine–Dalgarno sequence - Jin - 2006 - Molecular Microbiology - Wiley Online Library

Defining the anti-Shine-Dalgarno sequence interaction and quantifying its functional role in regulating translation efficiency | bioRxiv

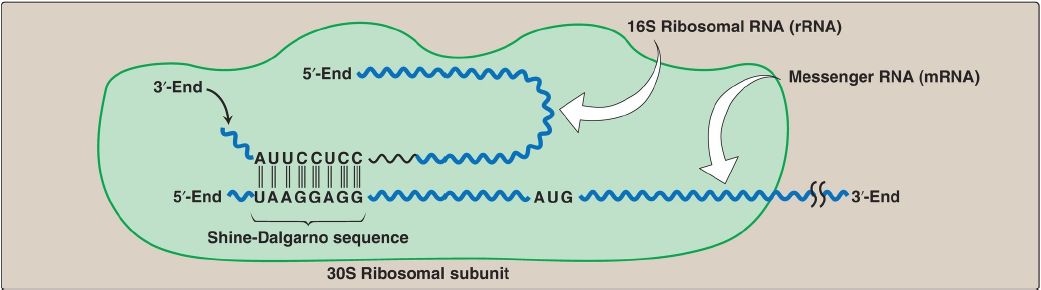
Shine-Dalgarno Motif Ribosome binding site located about 13 bases upstream of AUG start codon SD sequence is: 5'-AGGAGGU-3' Middle GGAG is more highly. - ppt download
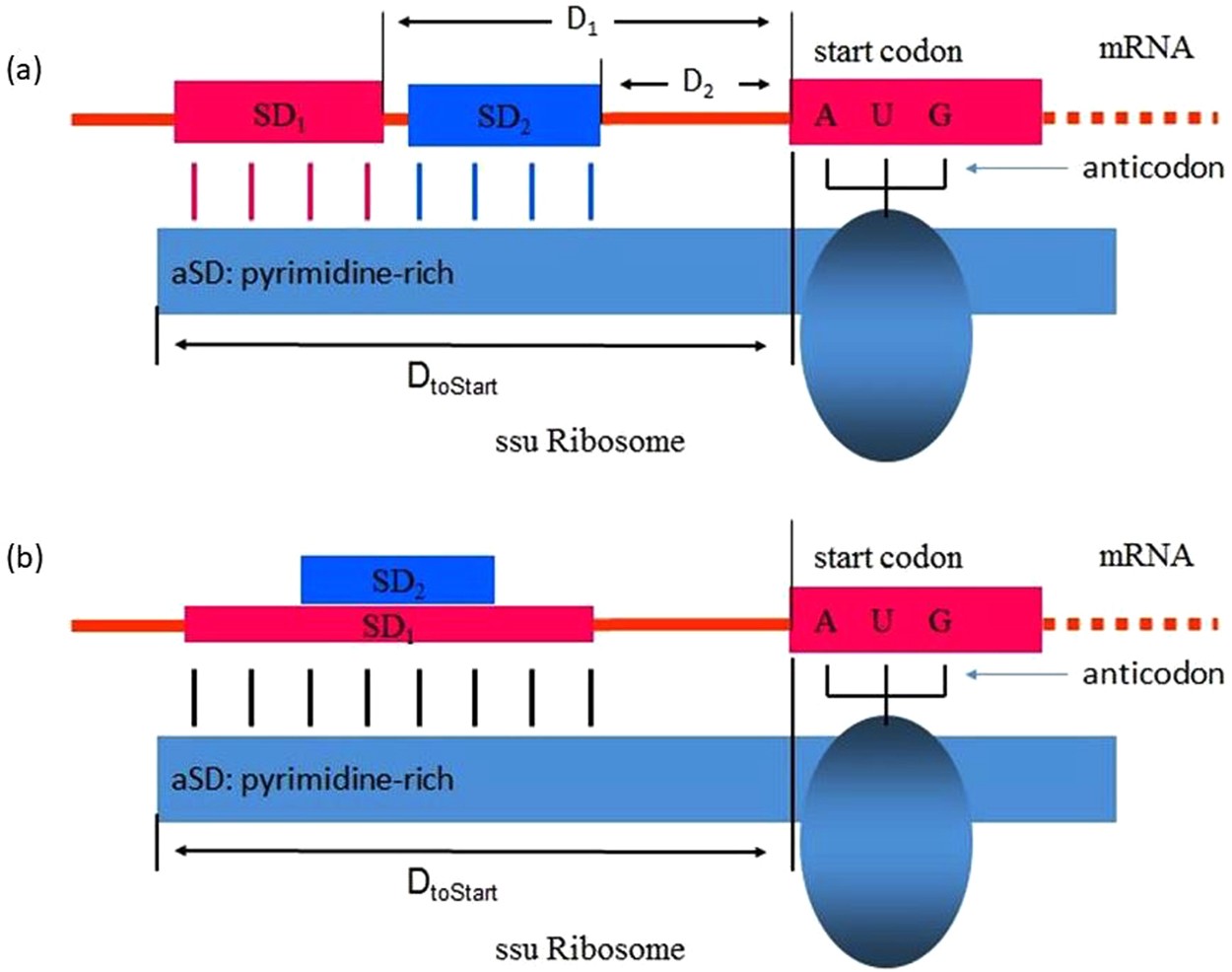
Elucidating the 16S rRNA 3′ boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data | Scientific Reports
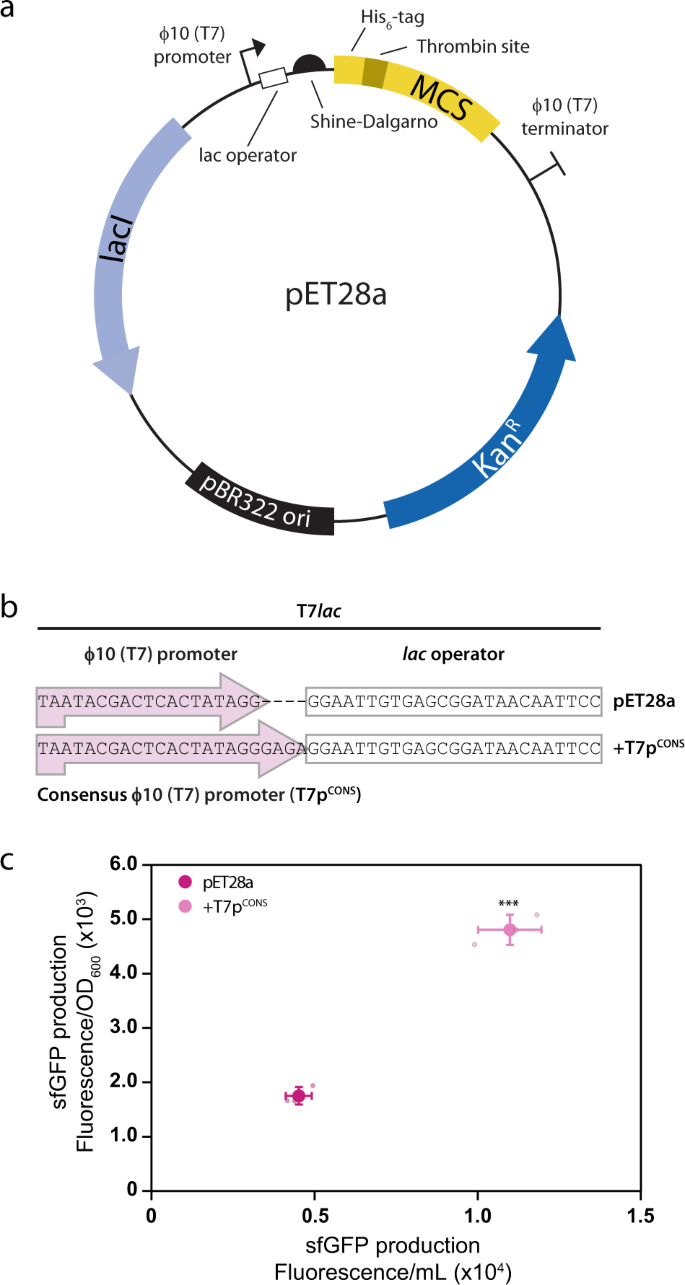
Improved designs for pET expression plasmids increase protein production yield in Escherichia coli | Communications Biology

Leveraging genome-wide datasets to quantify the functional role of the anti-Shine–Dalgarno sequence in regulating translation efficiency | Open Biology