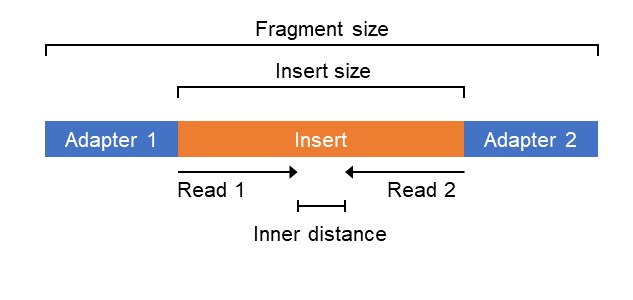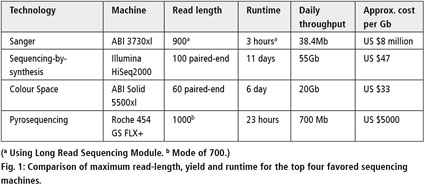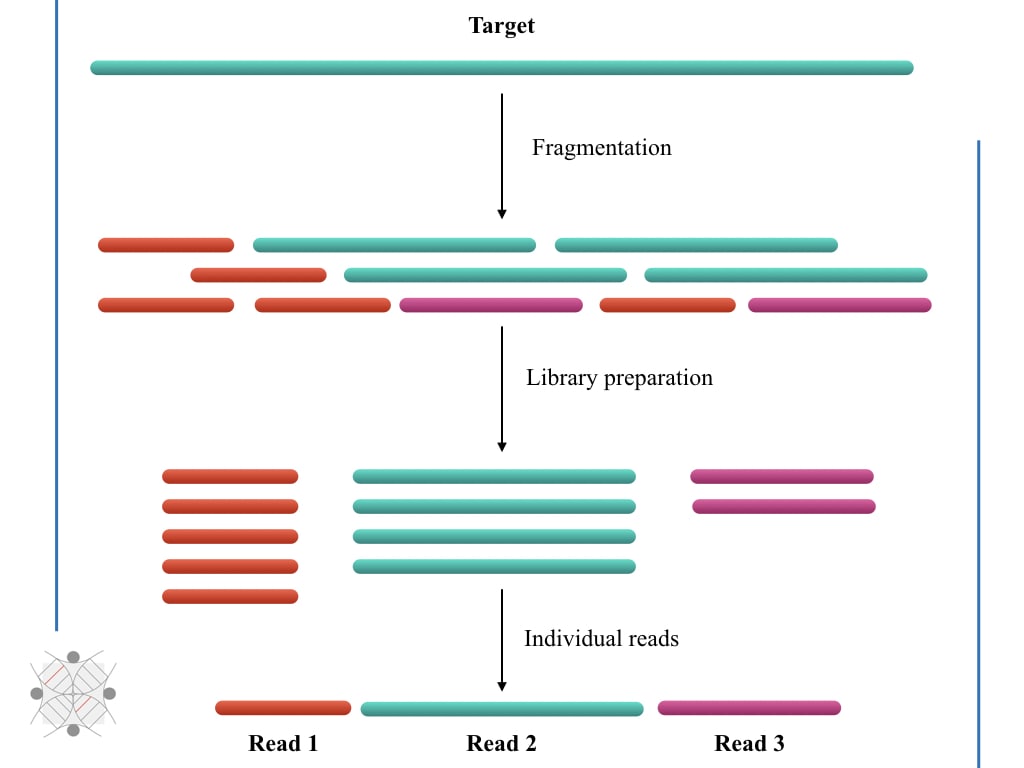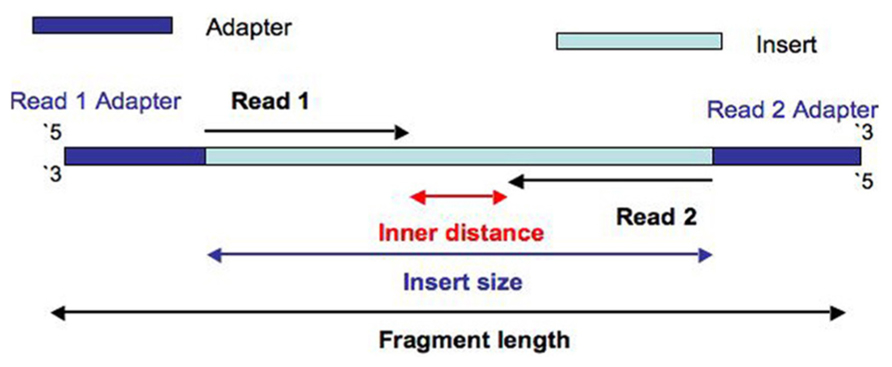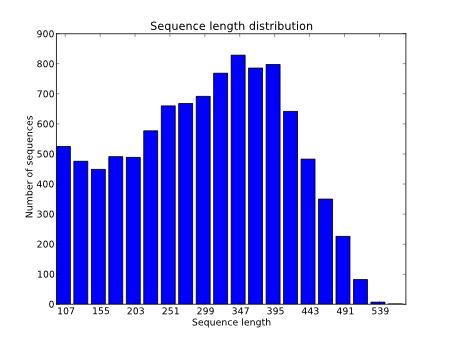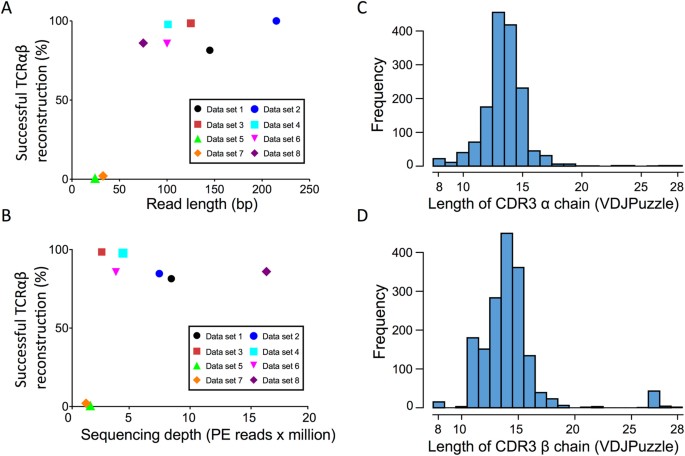
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

Oxford Nanopore Technology: A Promising Long-Read Sequencing Platform To Study Exon Connectivity and Characterize Isoforms of Complex Genes | Semantic Scholar
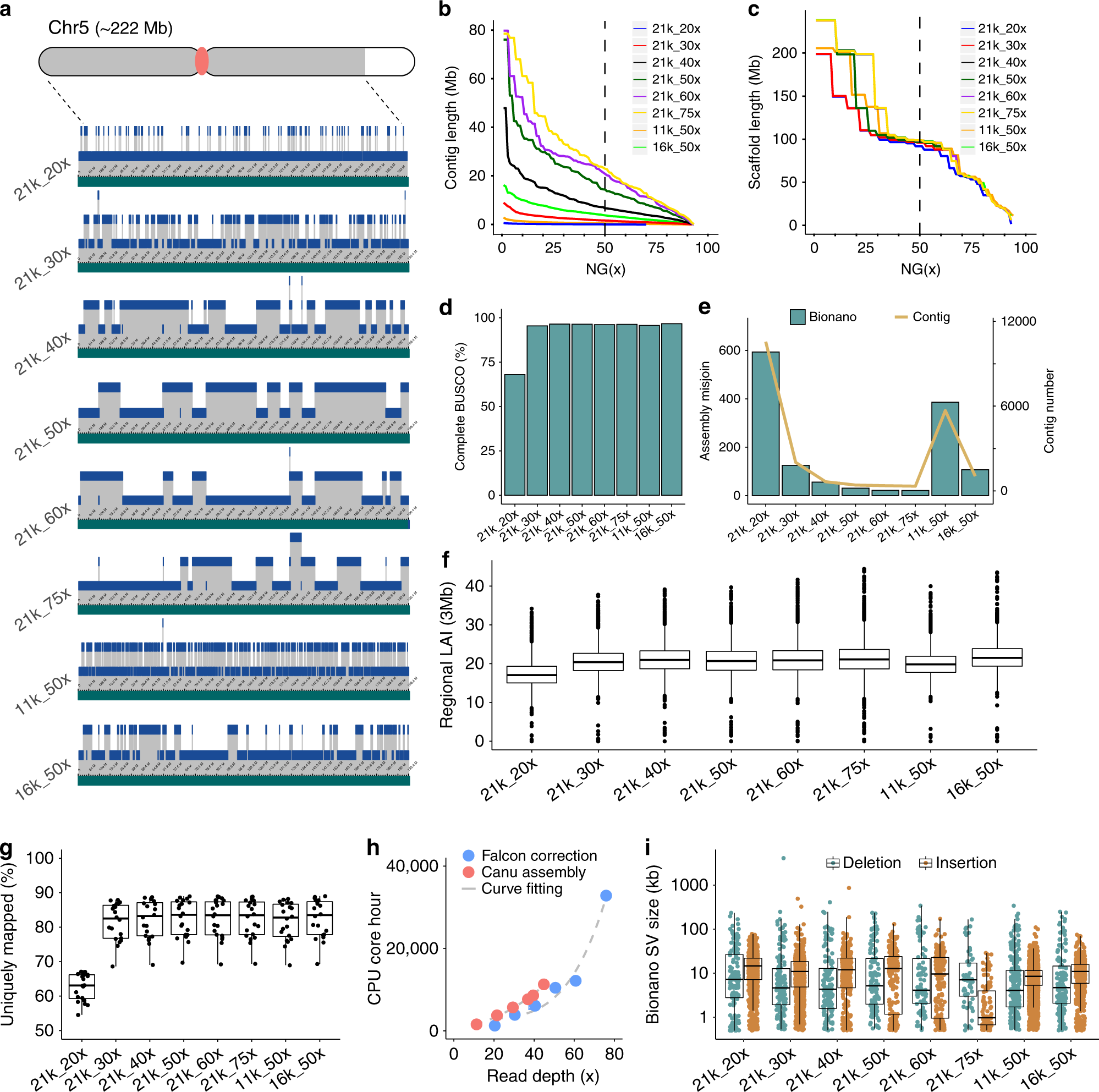
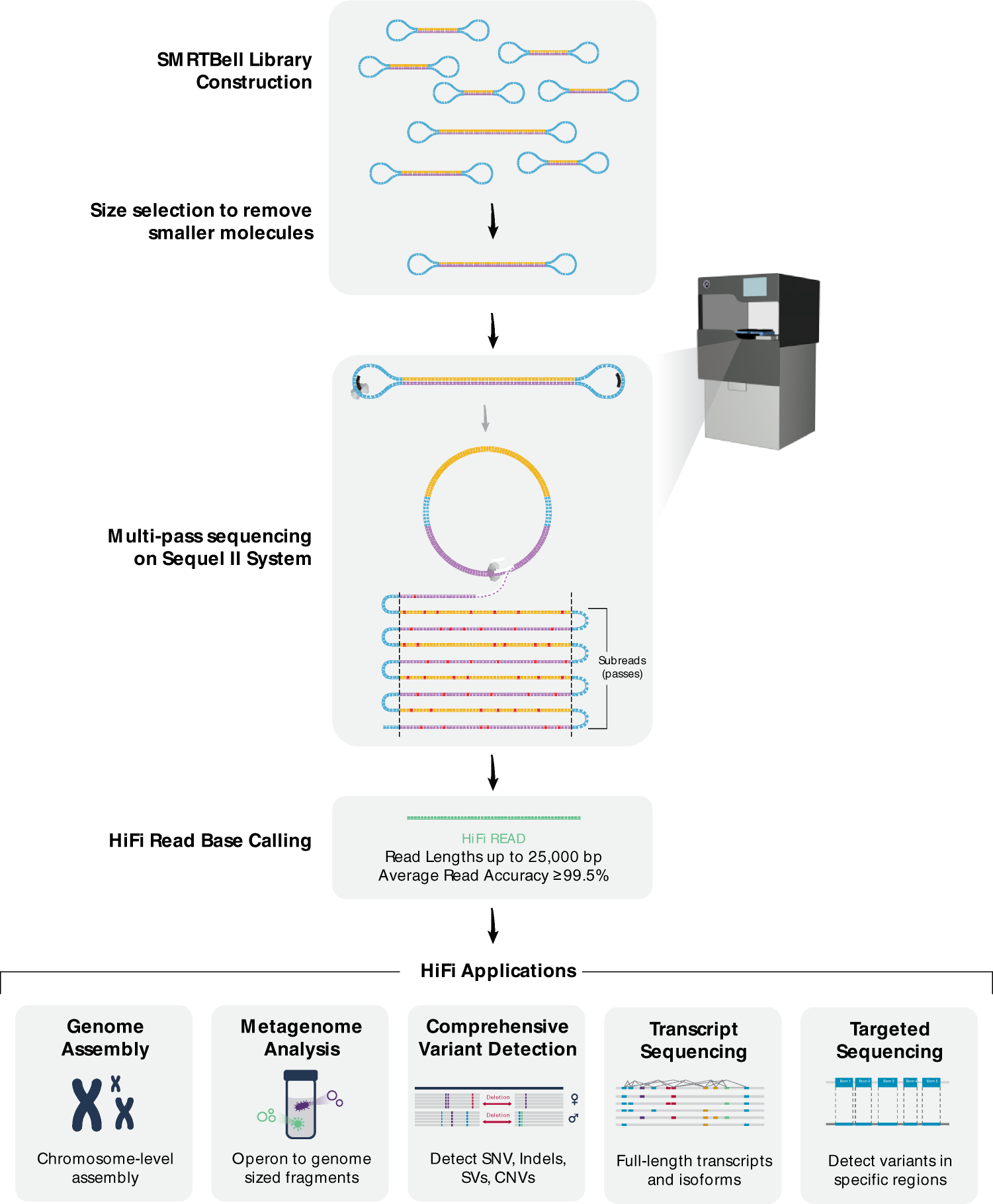
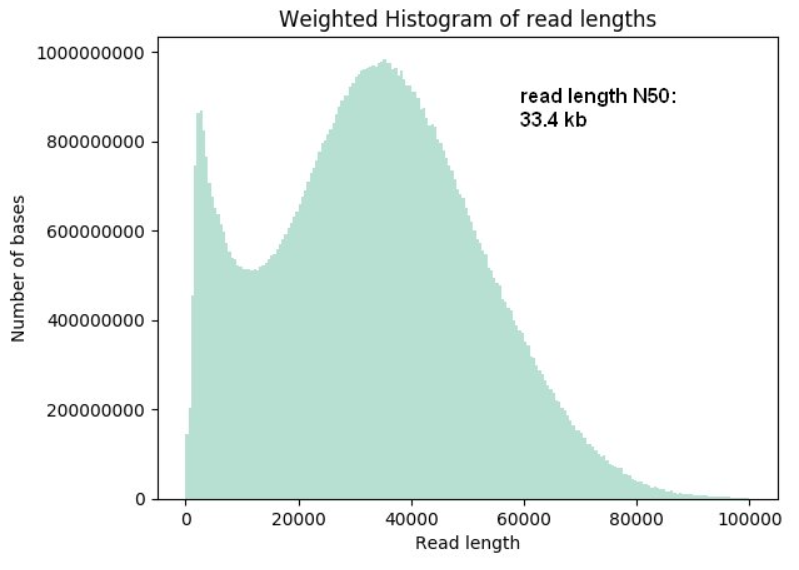
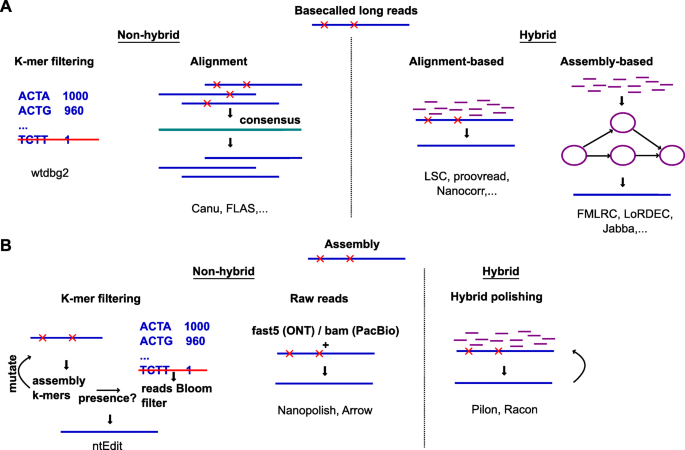
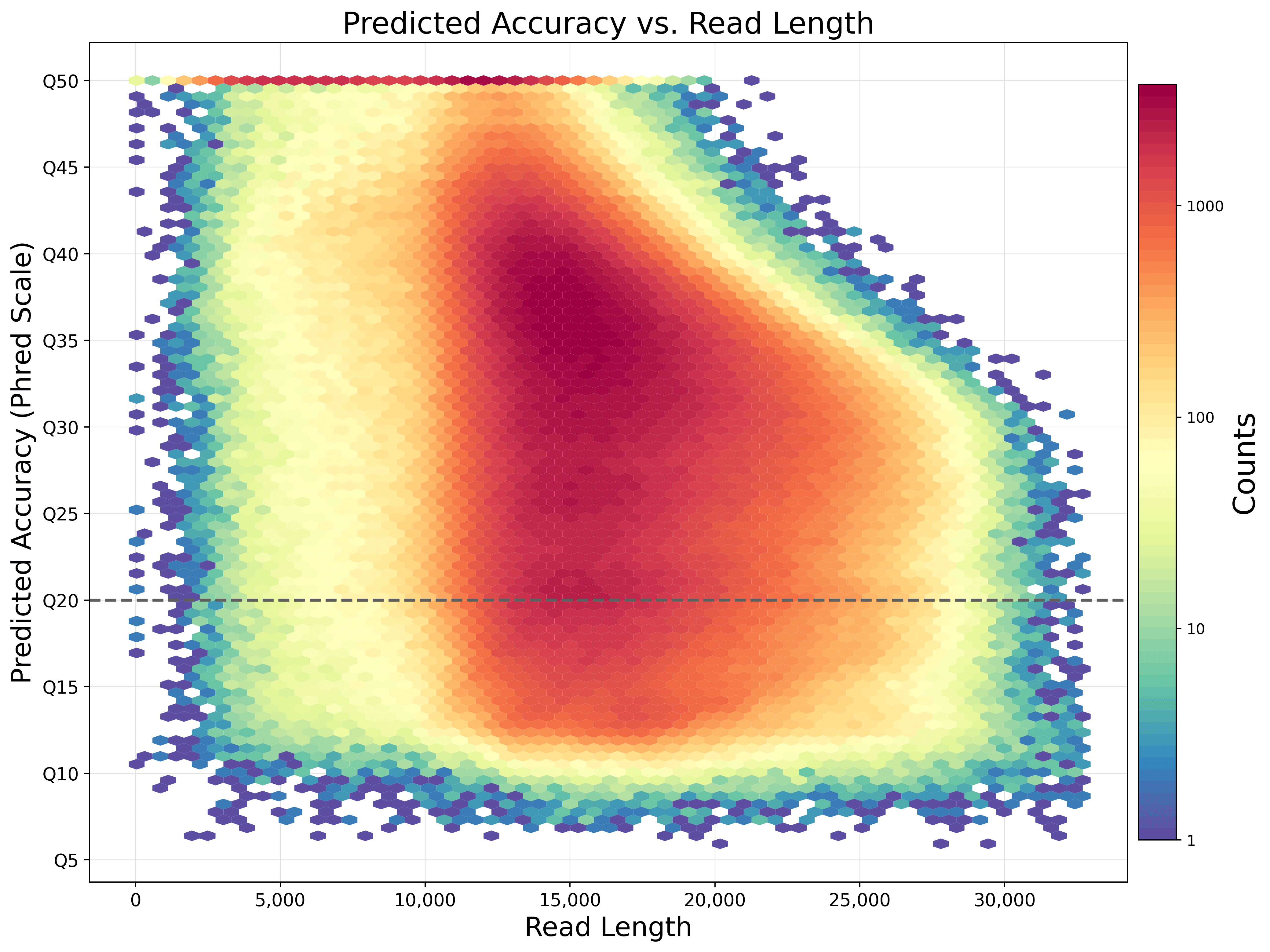
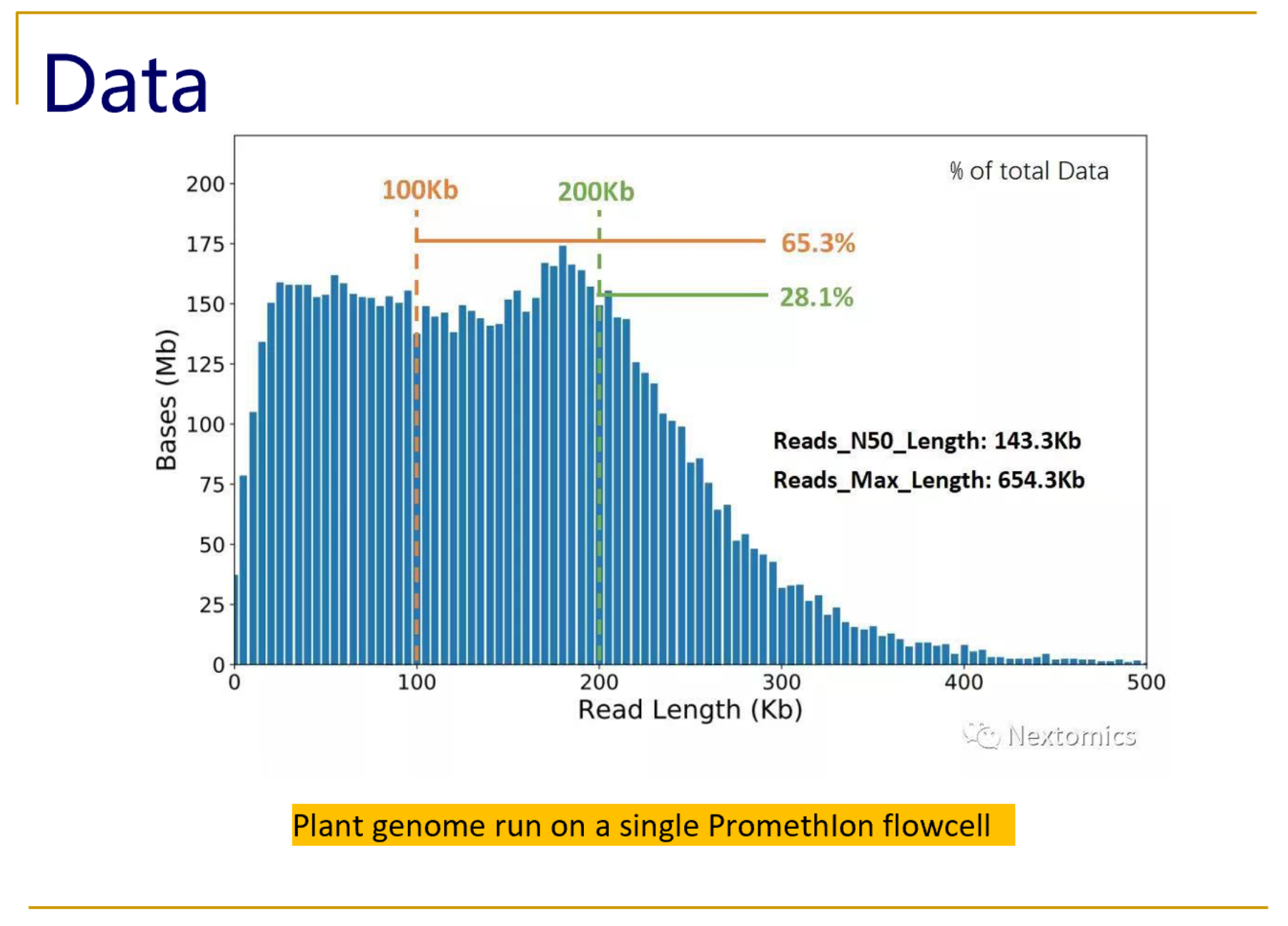
Effect of sequence depth and length in long-read assembly of the maize inbred NC358 | Nature Communications