![Navigating the amino acid sequence space between functional proteins using a deep learning framework [PeerJ] Navigating the amino acid sequence space between functional proteins using a deep learning framework [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/cs-684/1/fig-1-full.png)
Navigating the amino acid sequence space between functional proteins using a deep learning framework [PeerJ]

Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences | PNAS

MSA-Regularized Protein Sequence Transformer toward Predicting Genome-Wide Chemical-Protein Interactions: Application to GPCRome Deorphanization | Journal of Chemical Information and Modeling

Ultrafast end-to-end protein structure prediction enables high-throughput exploration of uncharacterized proteins | PNAS

Sequence similarity clustering of all MCE proteins. A representative... | Download Scientific Diagram
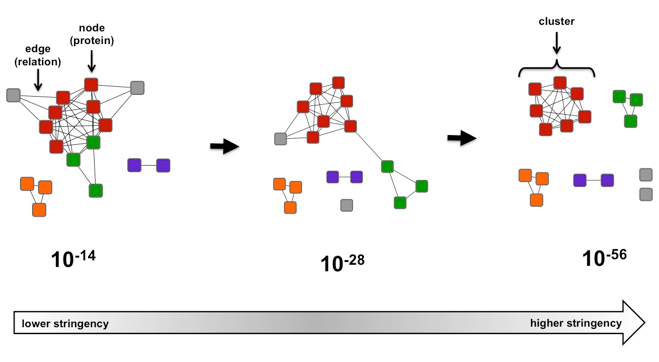
Analysis of plasmid genes by phylogenetic profiling and visualization of homology relationships using Blast2Network | BMC Bioinformatics | Full Text
GitHub - soedinglab/kClust: kClust is a fast and sensitive clustering method for the clustering of protein sequences. It is able to cluster large protein databases down to 20-30% sequence identity. kClust generates
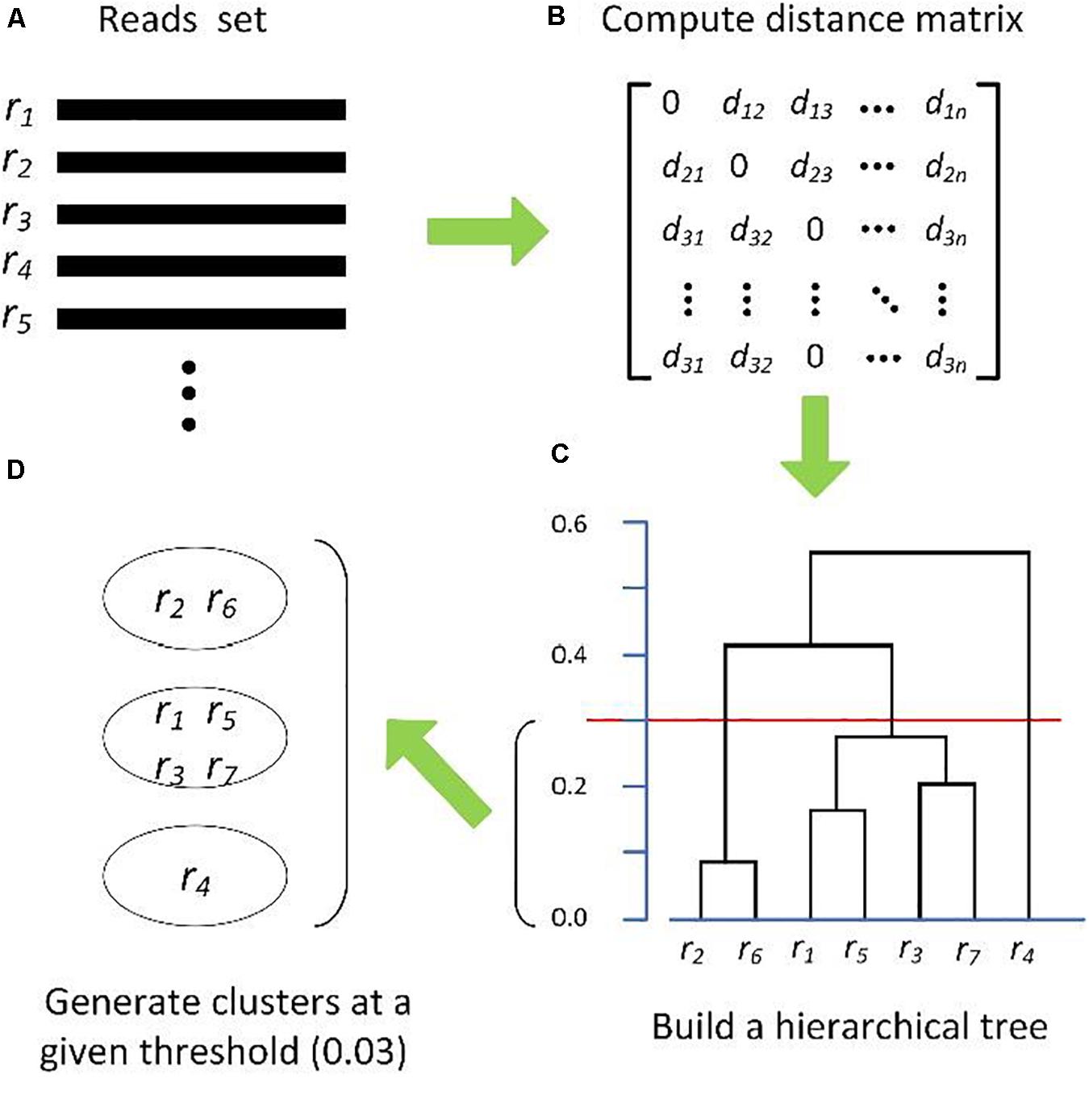
Frontiers | Comparison of Methods for Picking the Operational Taxonomic Units From Amplicon Sequences

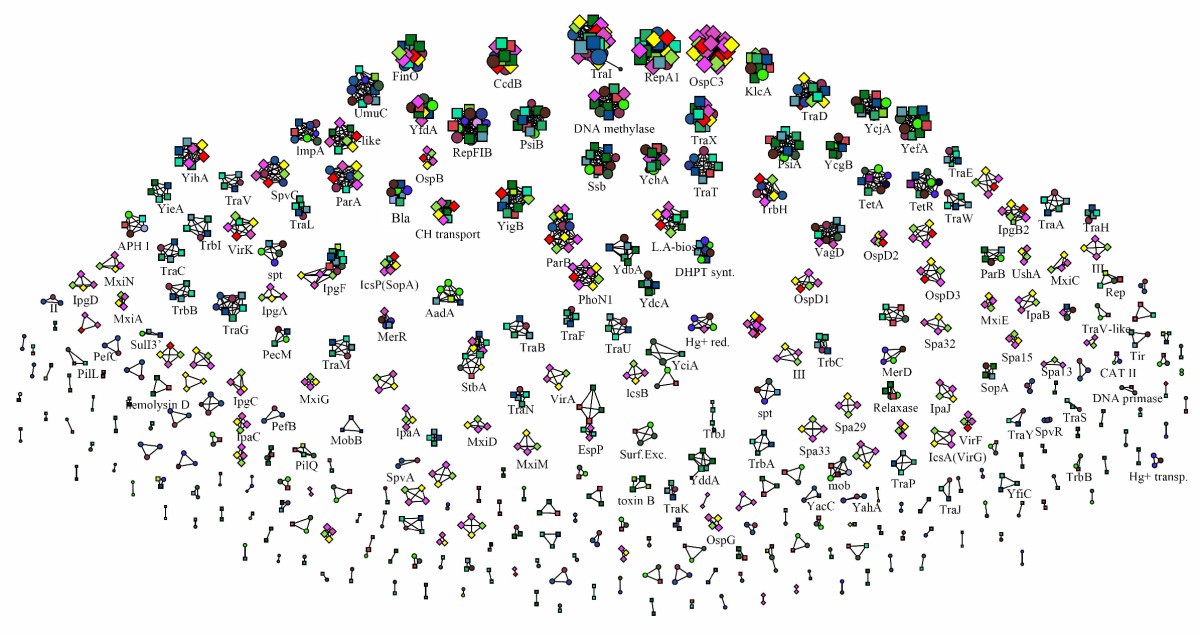
Visualizing and Clustering Protein Similarity Networks: Sequences, Structures, and Functions | Journal of Proteome Research

Partitioning clustering algorithms for protein sequence data sets – topic of research paper in Biological sciences. Download scholarly article PDF and read for free on CyberLeninka open science hub.
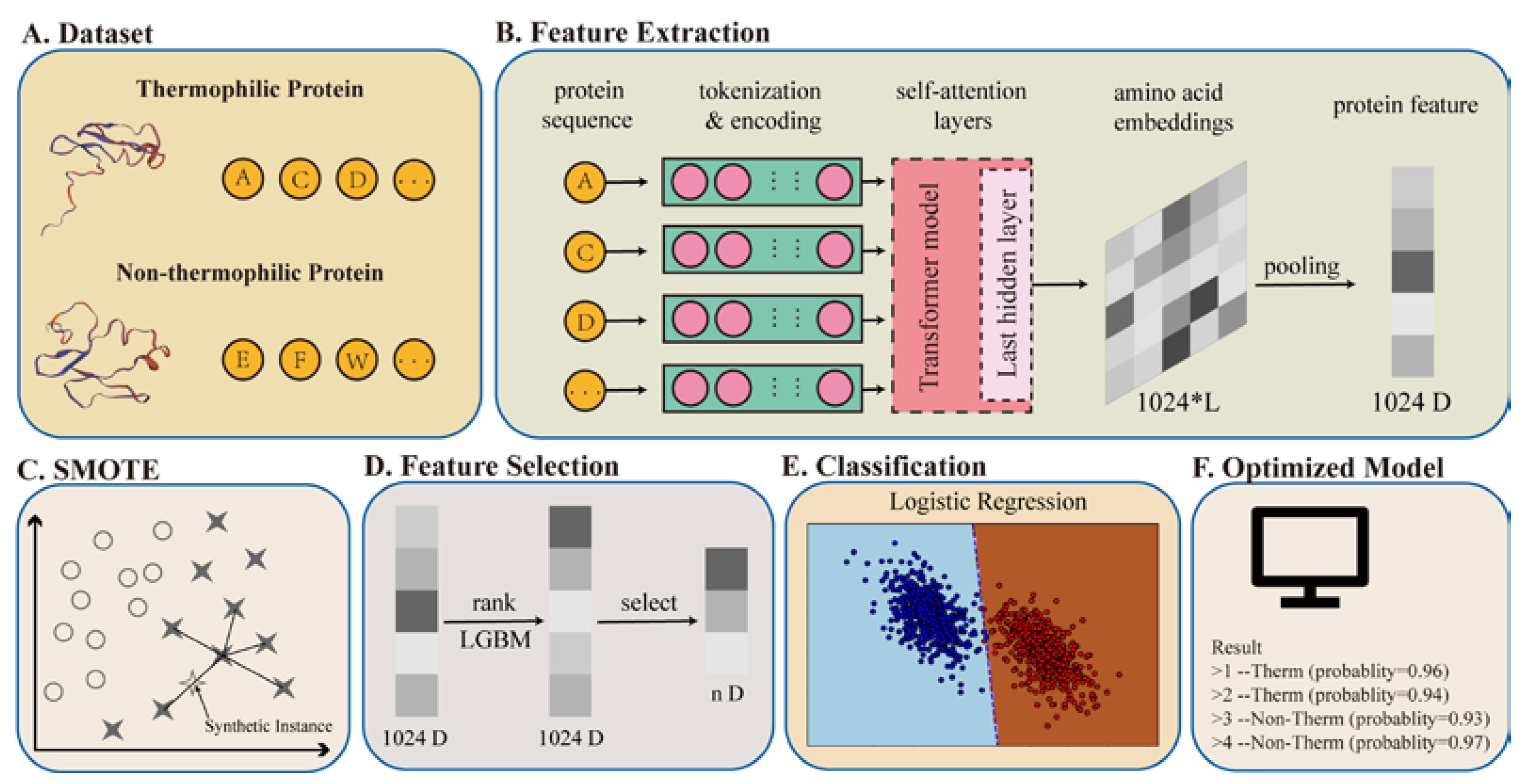
Applied Sciences | Free Full-Text | Identification of Thermophilic Proteins Based on Sequence-Based Bidirectional Representations from Transformer-Embedding Features

Development of a novel clustering tool for linear peptide sequences - Dhanda - 2018 - Immunology - Wiley Online Library

Claire McWhite on Twitter: "New preprint with @ProfMonaSingh. We present vcMSA, a totally new algorithm for multiple sequence alignment that's based on clustering protein language representations of amino acids. No gaps penalties,
![PDF] Minimum Spanning Tree-based Clustering Applied to Protein Sequences in Early Cancer Diagnosis | Semantic Scholar PDF] Minimum Spanning Tree-based Clustering Applied to Protein Sequences in Early Cancer Diagnosis | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e887dab367f4c77f8b70f7d7a90b8fe77747a9bf/4-Figure1-1.png)
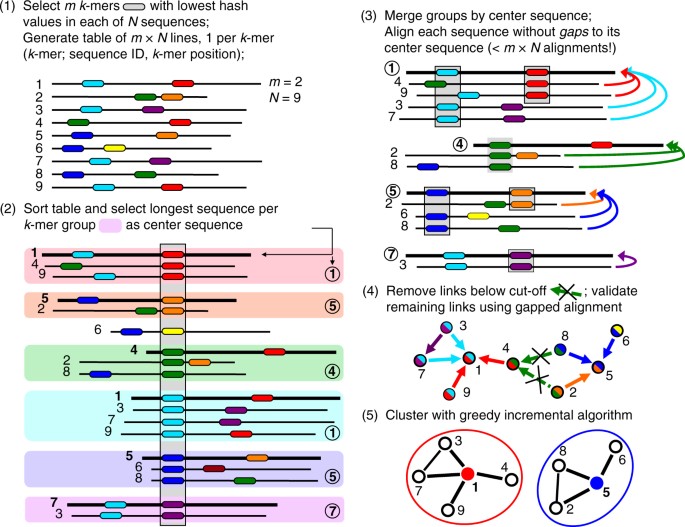



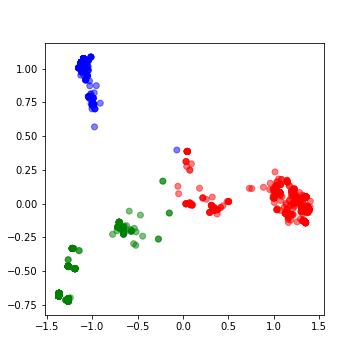
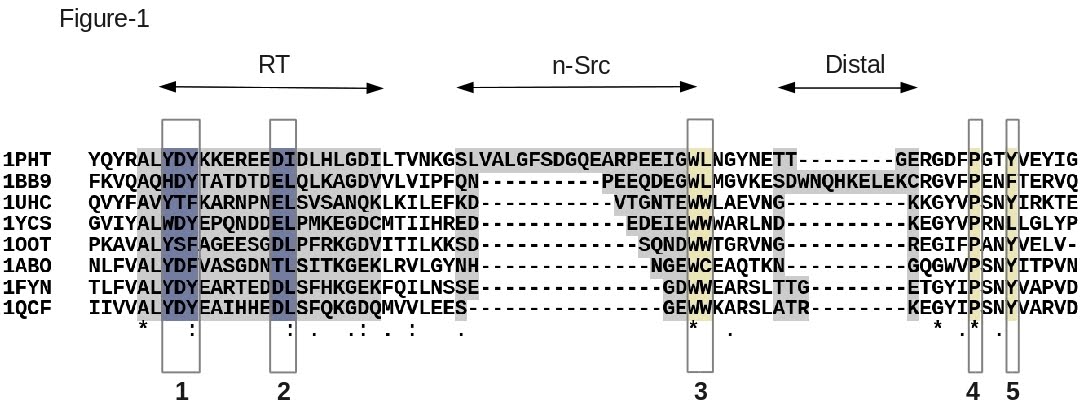
![Help [PIR - Protein Information Resource] Help [PIR - Protein Information Resource]](https://proteininformationresource.org/pirwww/images/creationpirsf.jpg)