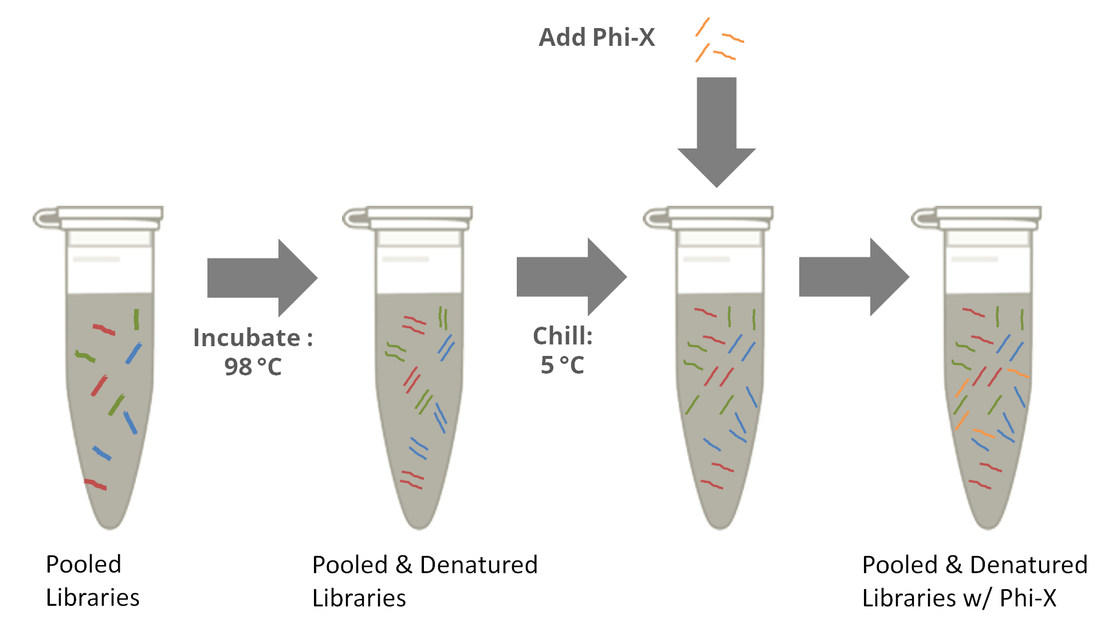
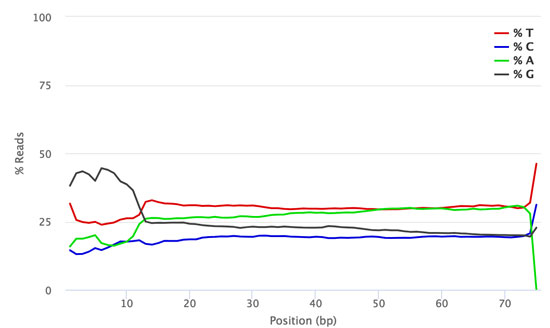
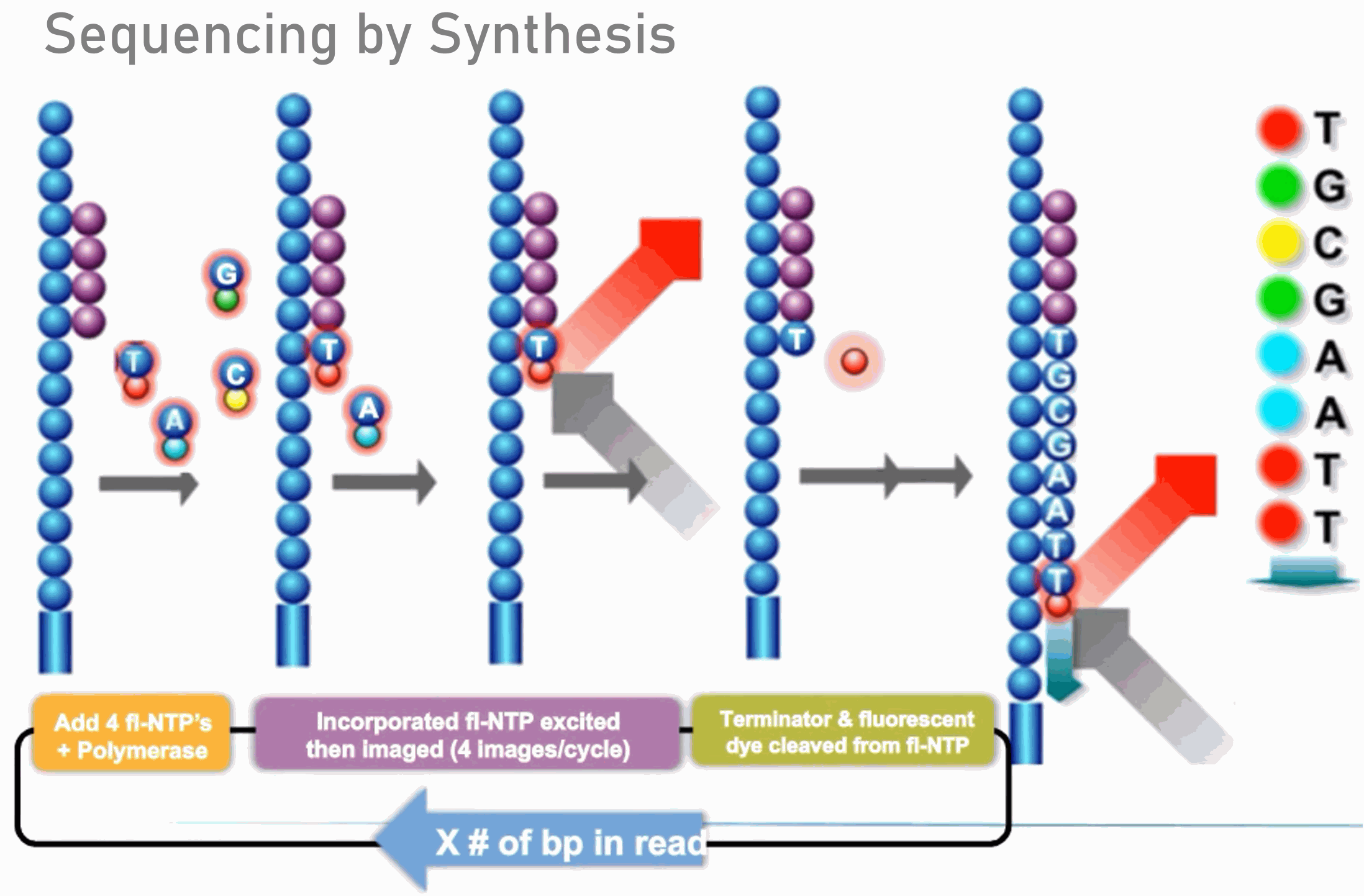
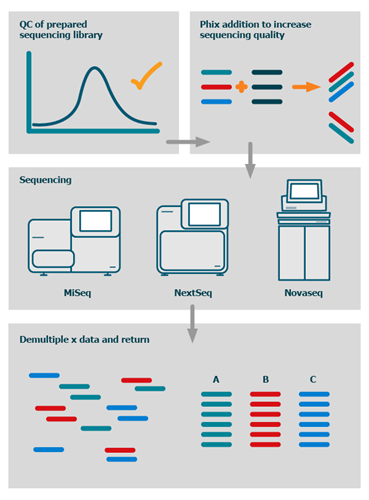
How much PhiX spike in is recommended when sequencing low diversity libraries on Illumina platforms? - Illumina Knowledge
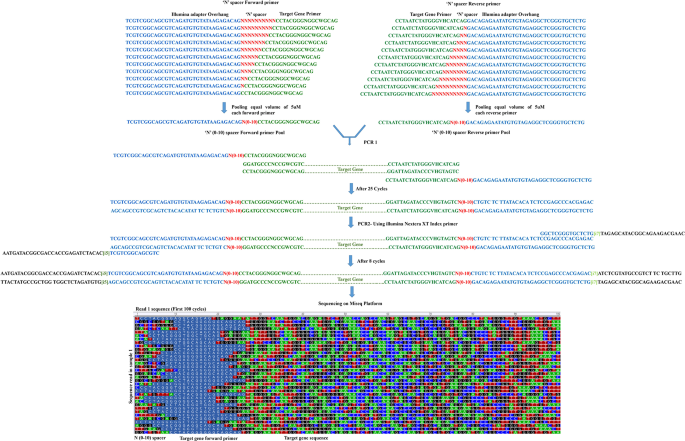
High-quality single amplicon sequencing method for illumina MiSeq platform using pool of 'N' (0–10) spacer-linked target specific primers without PhiX spike-in | BMC Genomics | Full Text