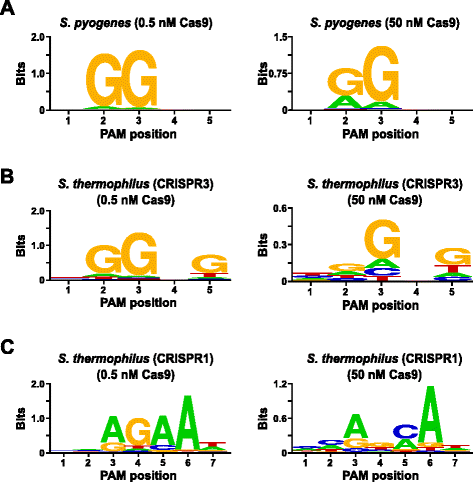
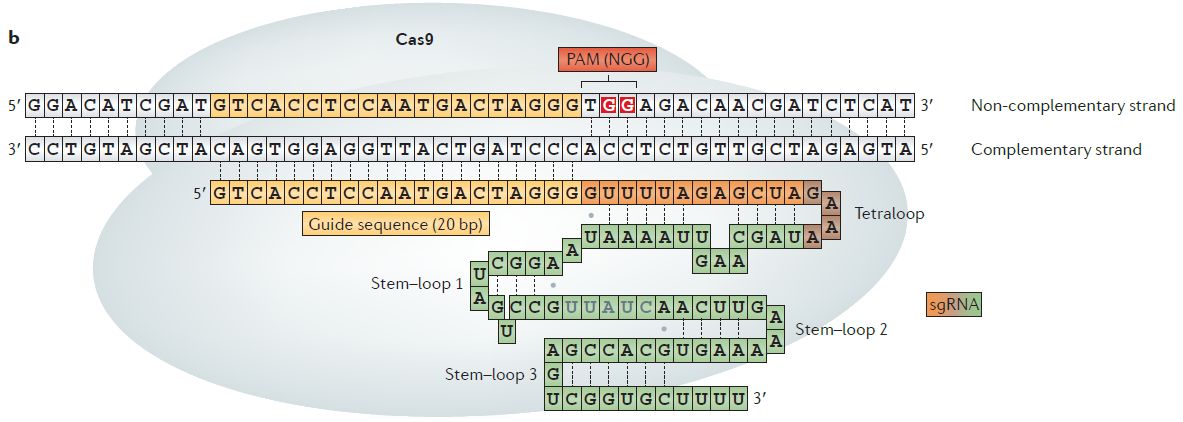
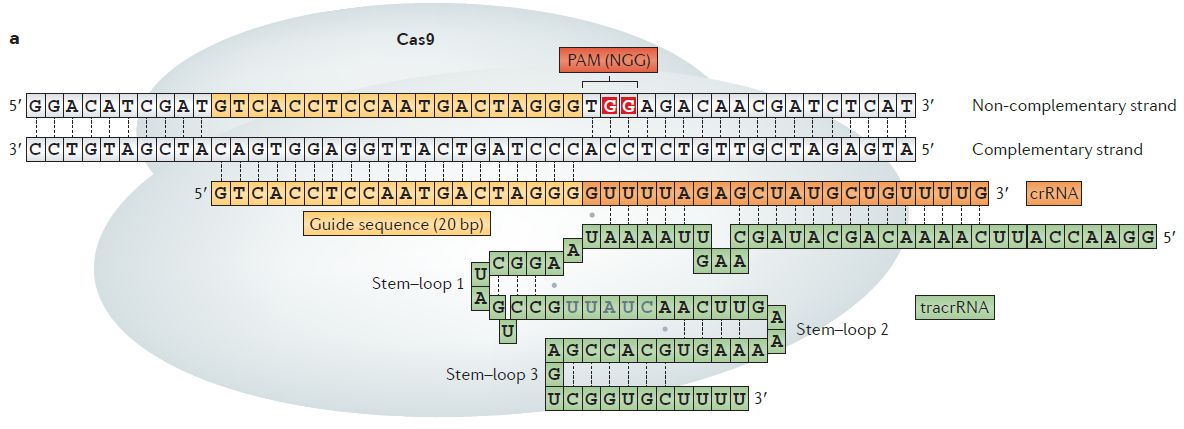
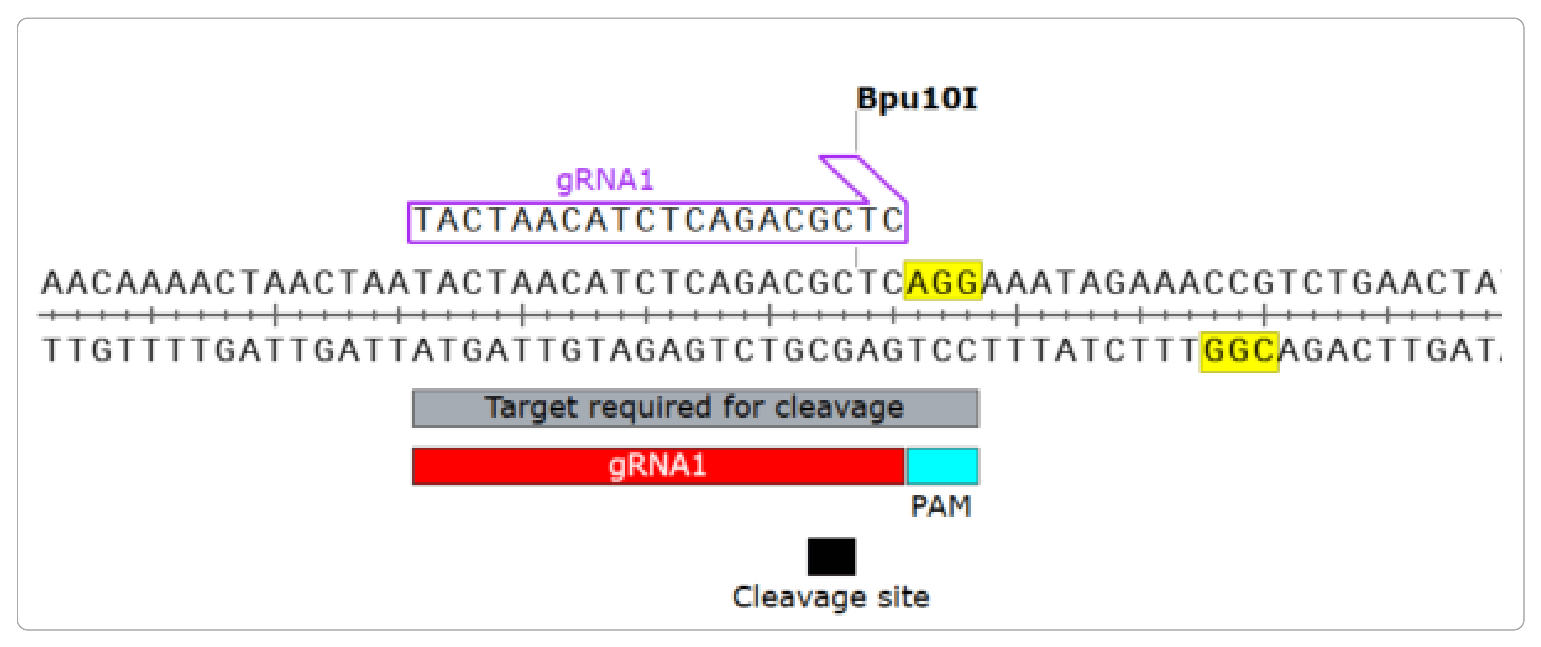
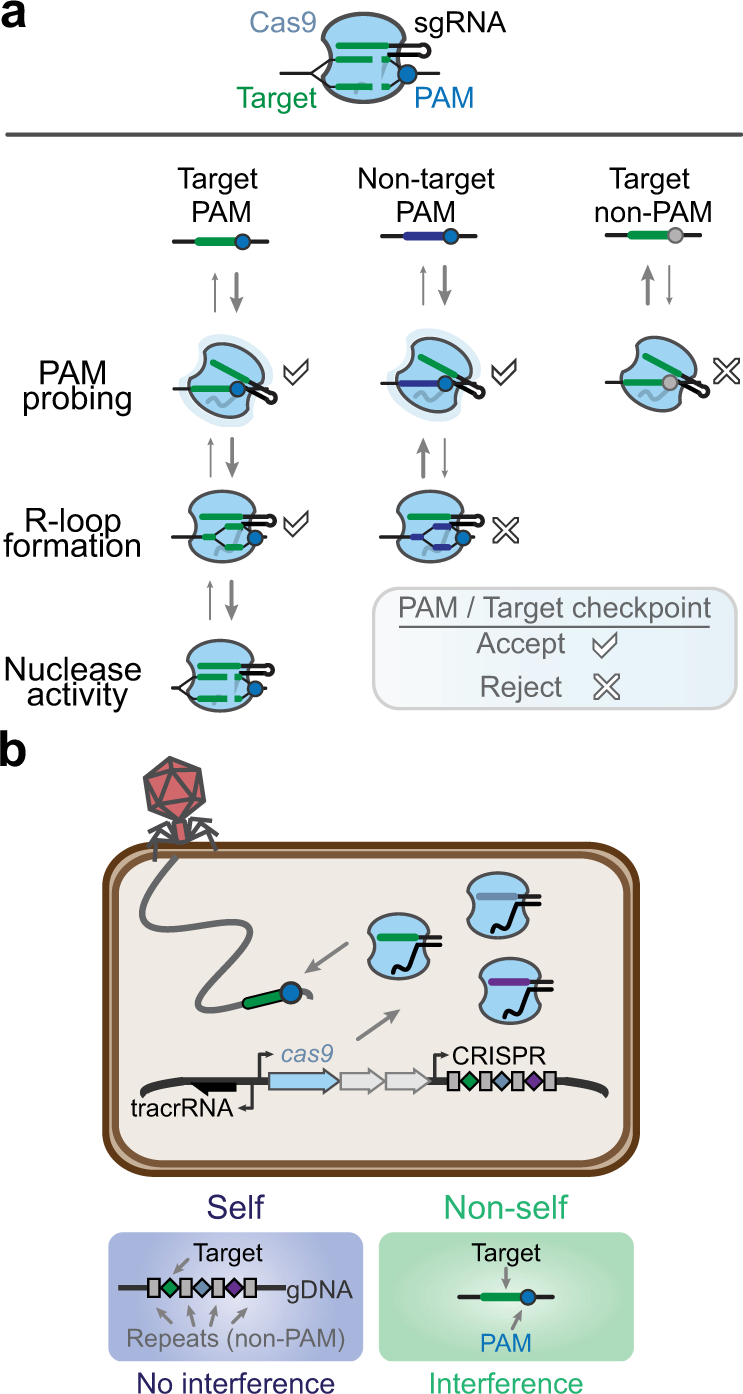
Comprehensive PAM prediction for CRISPR-Cas systems reveals evidence for spacer sharing, preferred strand targeting and conserved links with CRISPR repeats | bioRxiv
CRISPR Knock-in Designer: Automatic Oligonucleotide Design Software to Introduce Point Mutations by Gene Editing Methods

A highly reliable PAM prediction algorithm a Schematic description of... | Download Scientific Diagram
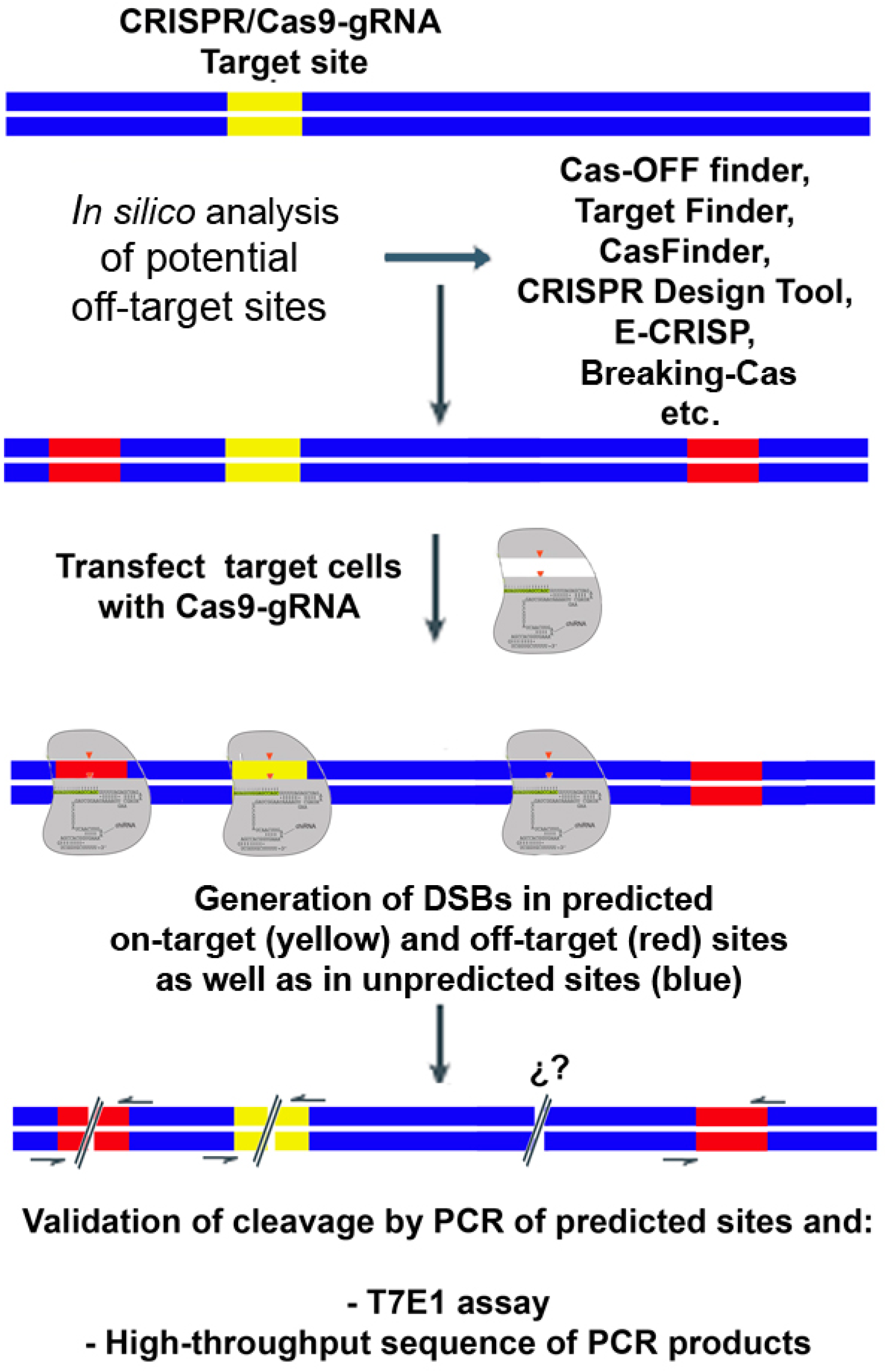
IJMS | Free Full-Text | Biased and Unbiased Methods for the Detection of Off-Target Cleavage by CRISPR/Cas9: An Overview

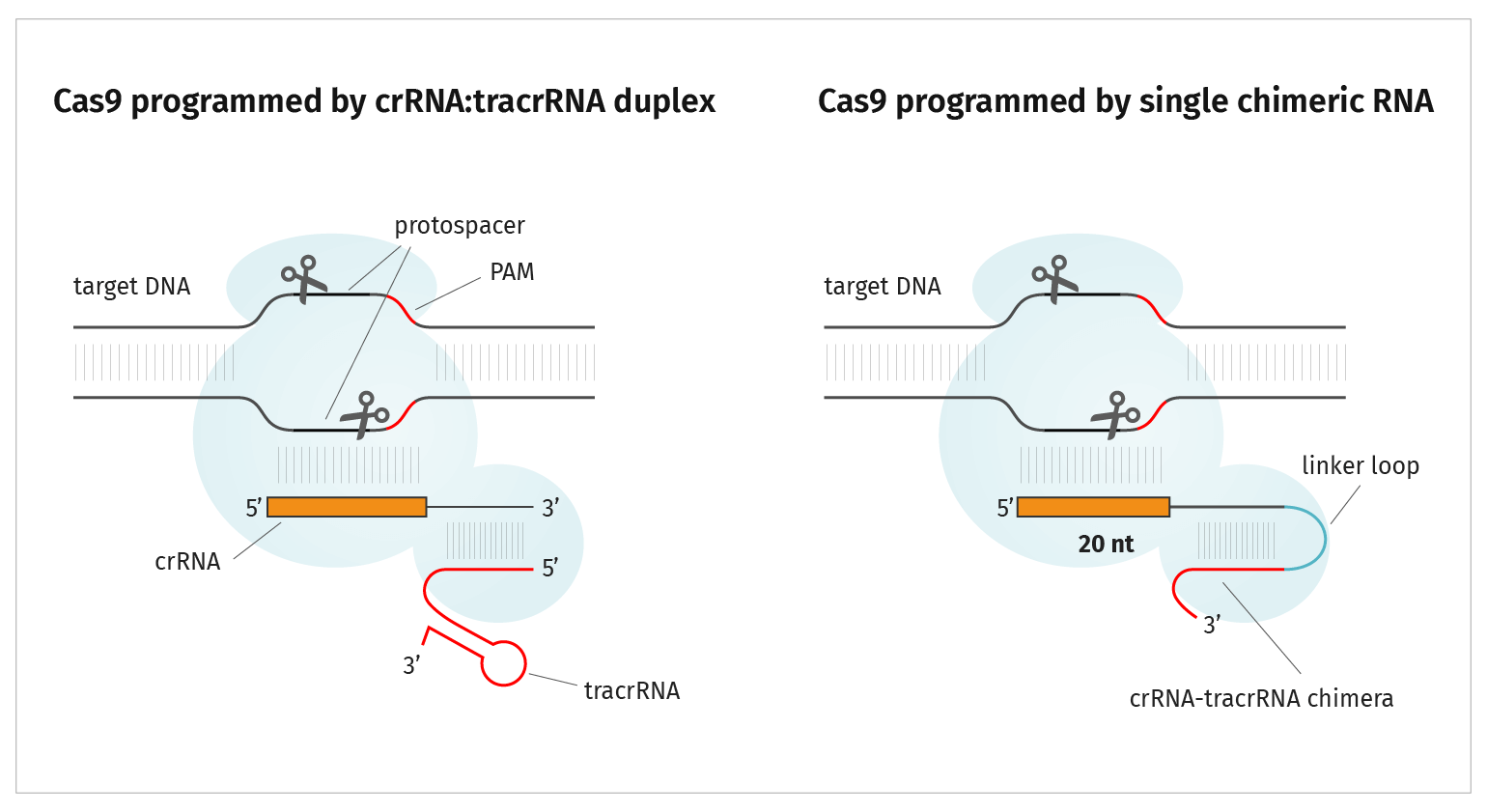
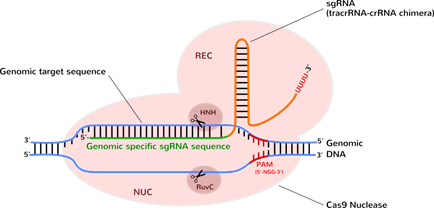
Identification of functional PAM sequences for CRISPR-interference.... | Download Scientific Diagram
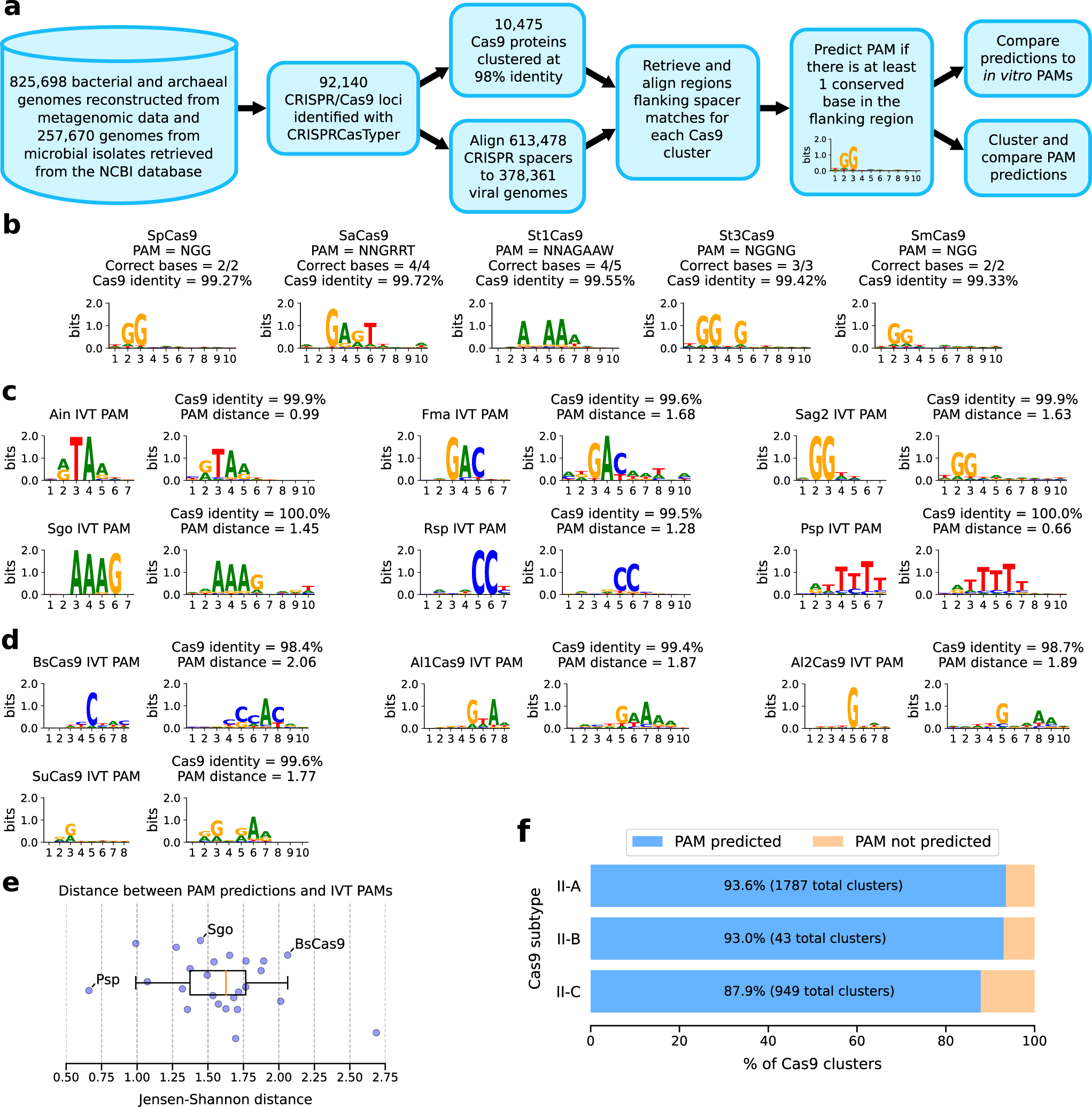
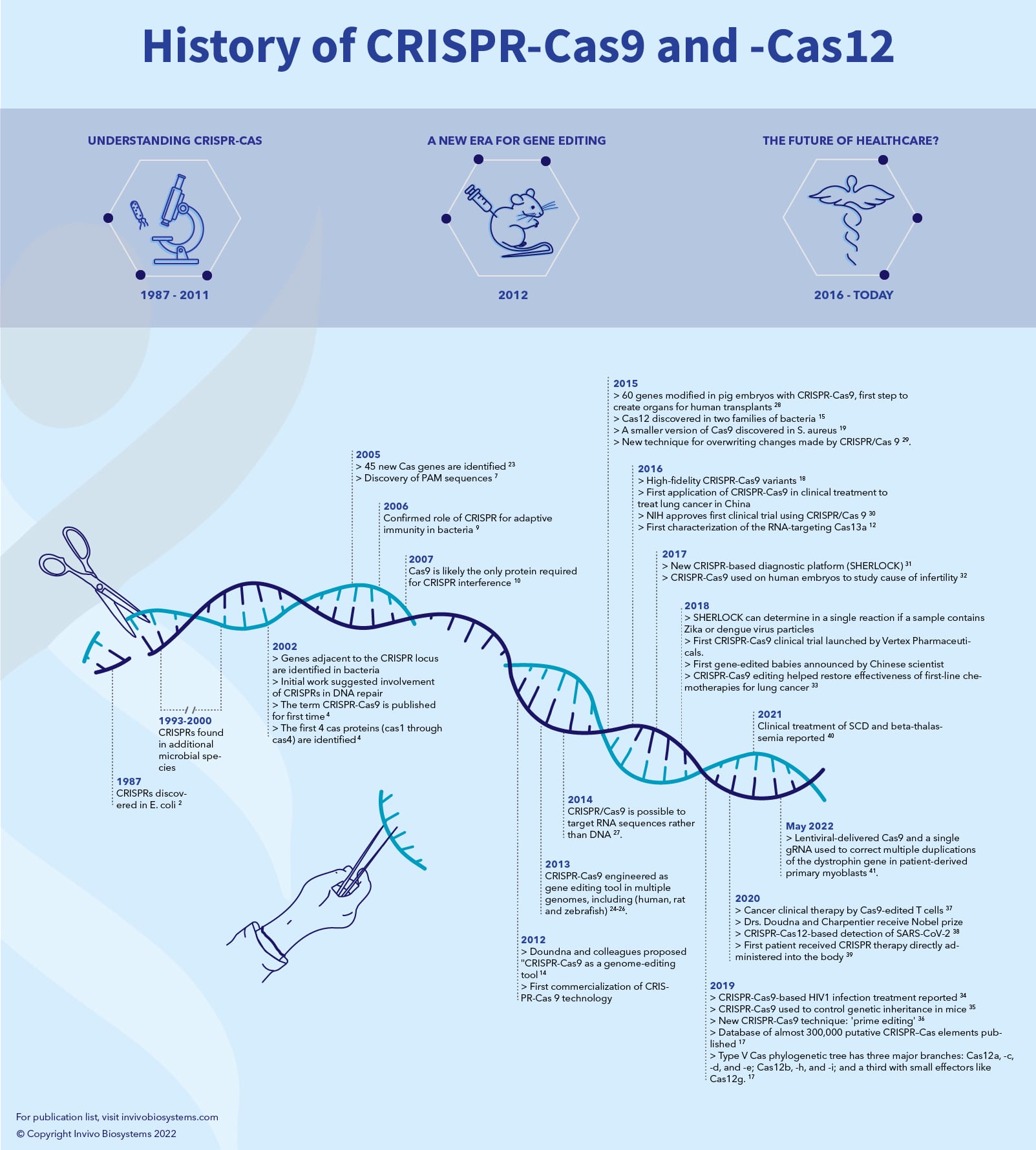
Automated identification of sequence-tailored Cas9 proteins using massive metagenomic data | Nature Communications

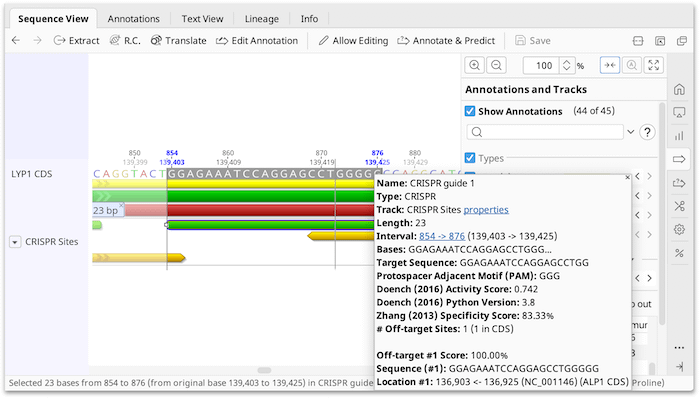
Nuclease Target Site Selection for Maximizing On-target Activity and Minimizing Off-target Effects in Genome Editing: Molecular Therapy

In Silico Processing of the Complete CRISPR‐Cas Spacer Space for Identification of PAM Sequences - Mendoza - 2018 - Biotechnology Journal - Wiley Online Library

Comprehensive PAM prediction for CRISPR-Cas systems reveals evidence for spacer sharing, preferred strand targeting and conserved links with CRISPR repeats | bioRxiv