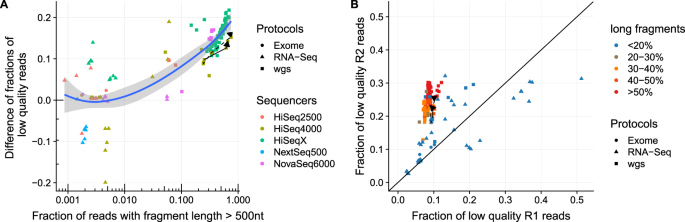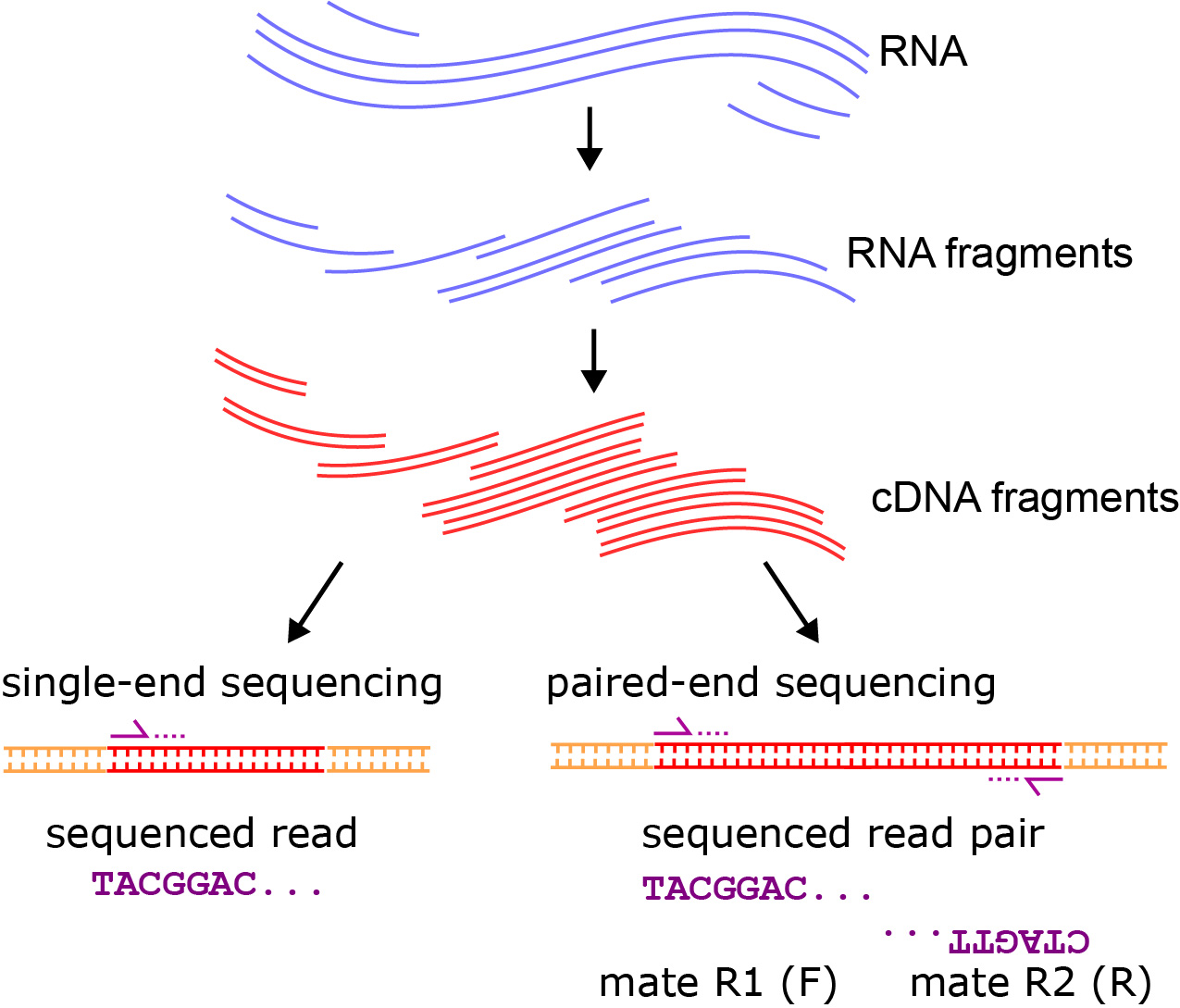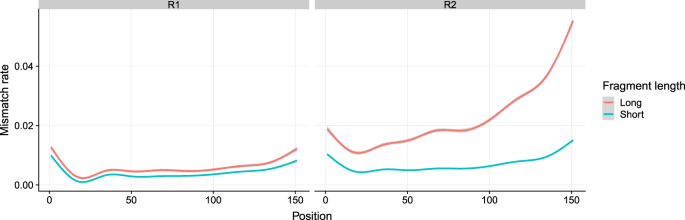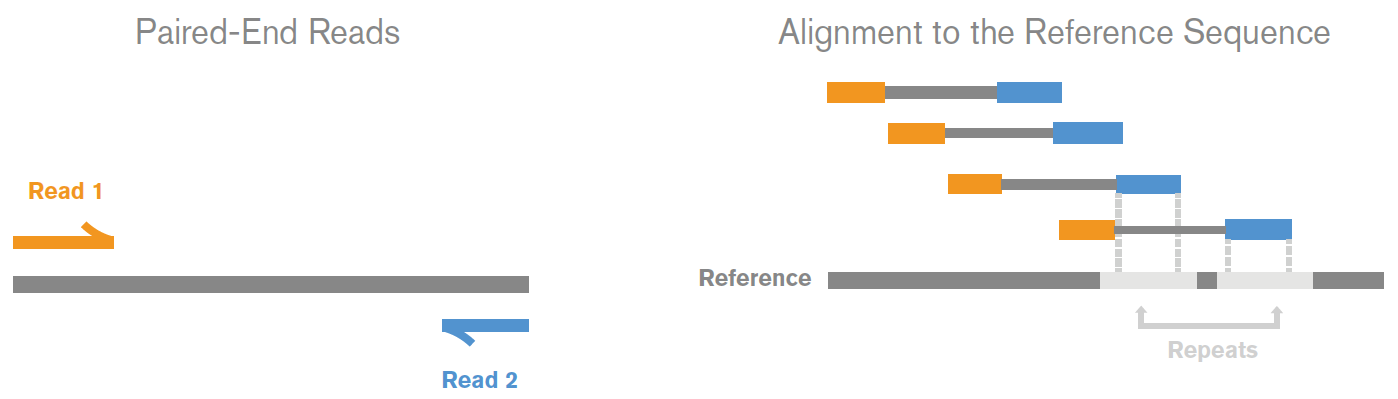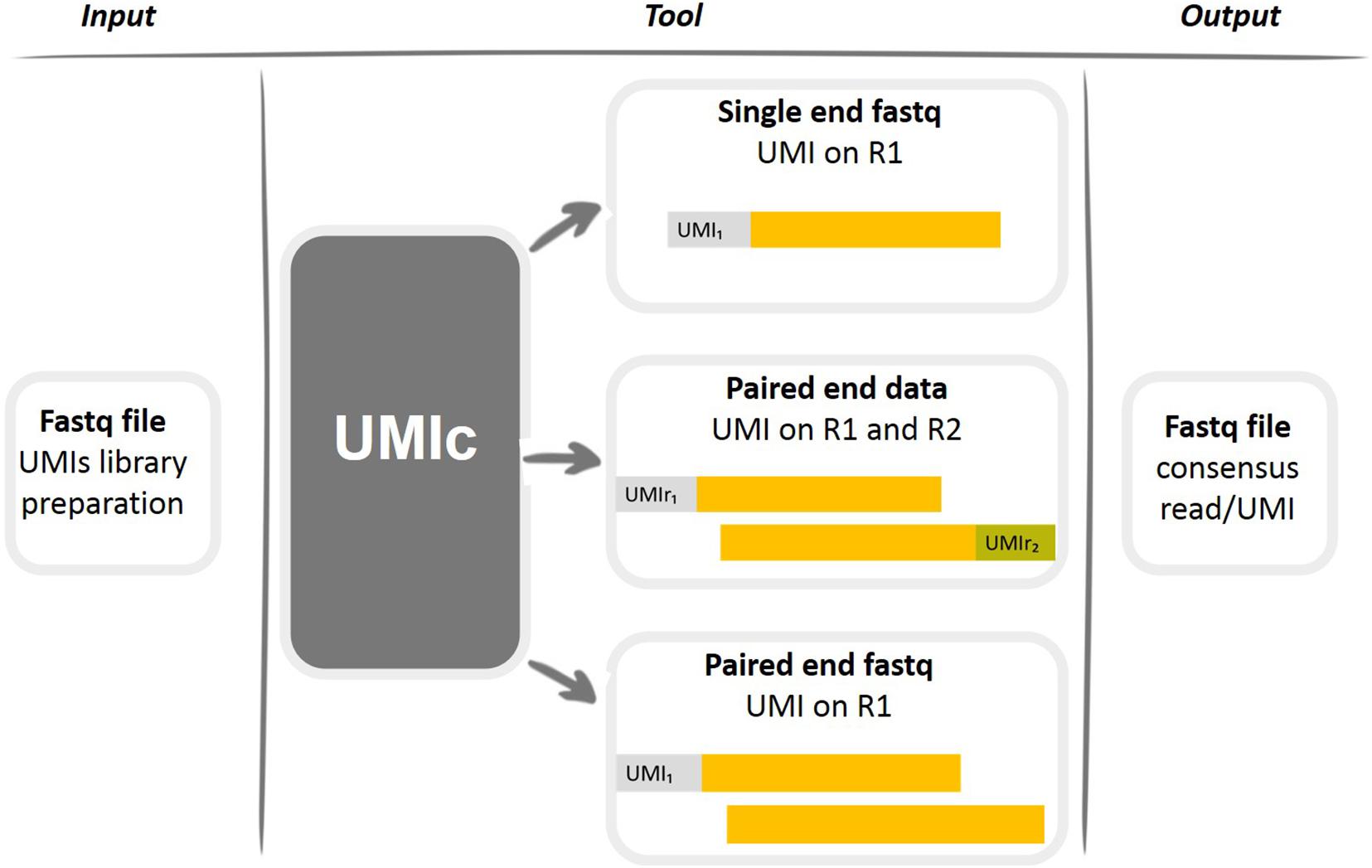
NGmerge: merging paired-end reads via novel empirically-derived models of sequencing errors | BMC Bioinformatics | Full Text

Mismatch rates detected from mapping reads to reference genome. Reads... | Download Scientific Diagram
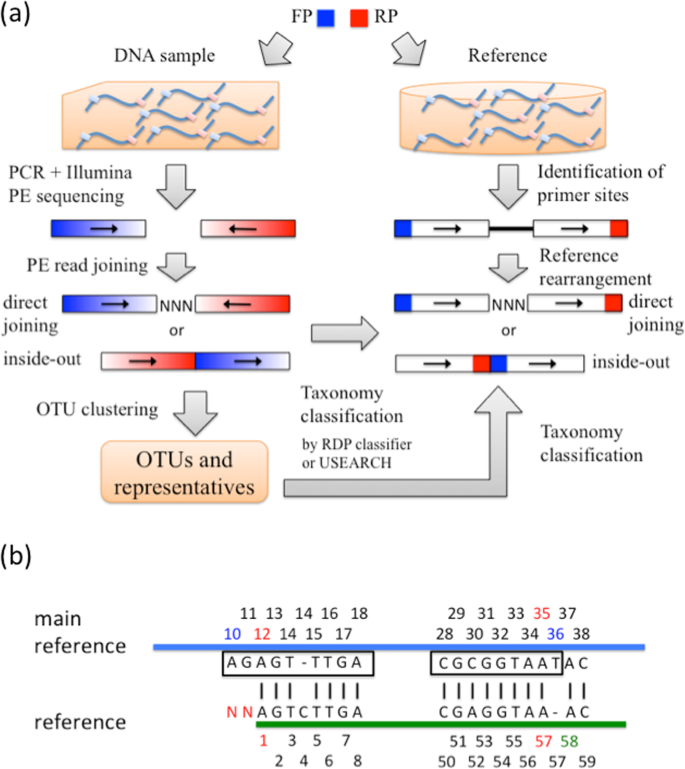
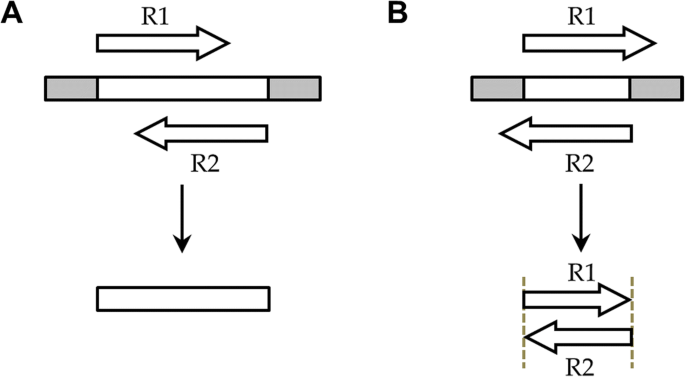
Joining Illumina paired-end reads for classifying phylogenetic marker sequences | BMC Bioinformatics | Full Text

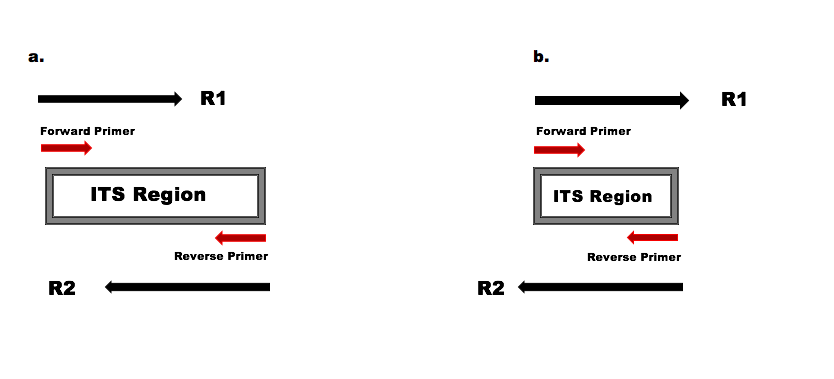
The direction and positional order of the paired-end reads (R1–R2). If... | Download Scientific Diagram

How to quantify the overlapping reads in paired-end DNA sequencing to check the sequencing efficiency
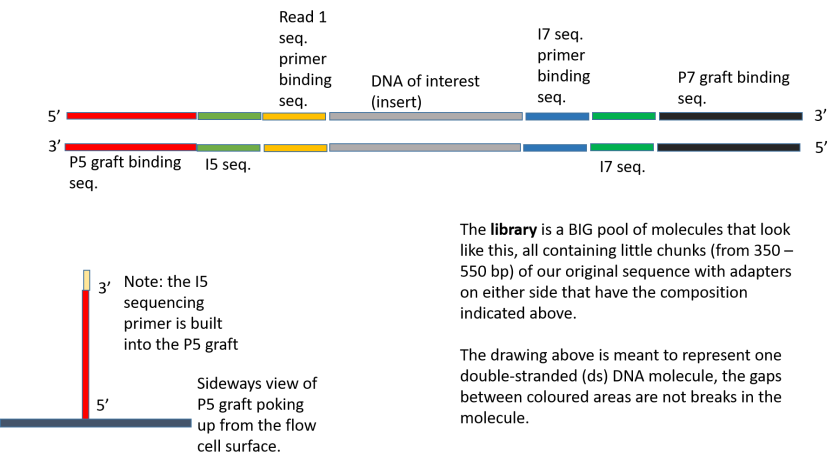
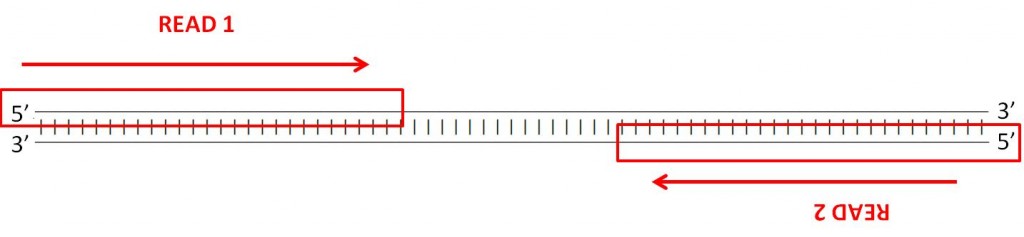
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics
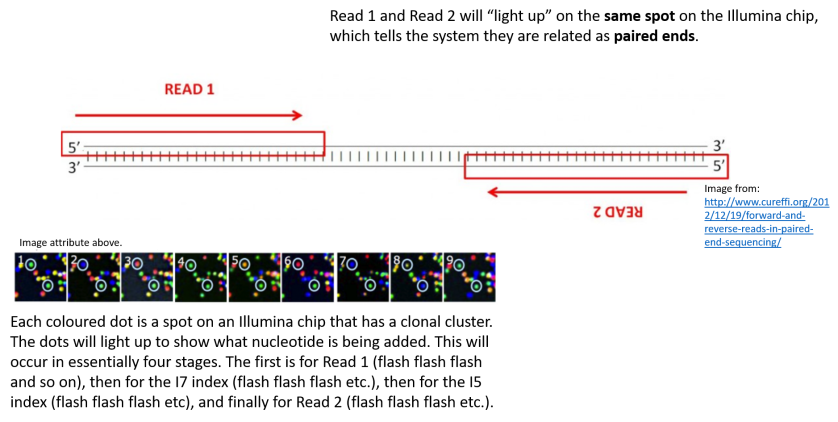
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics