Deep learning on computational biology and bioinformatics tutorial: from DNA to protein folding and alphafold2 | AI Summer
Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning

Protein Sequence Classification. A case study on Pfam dataset to… | by Ronak Vijay | Towards Data Science

A Convolutional Neural Network Using Dinucleotide One-hot Encoder for identifying DNA N6-Methyladenine Sites in the Rice Genome - ScienceDirect

Neural networks to learn protein sequence–function relationships from deep mutational scanning data | PNAS
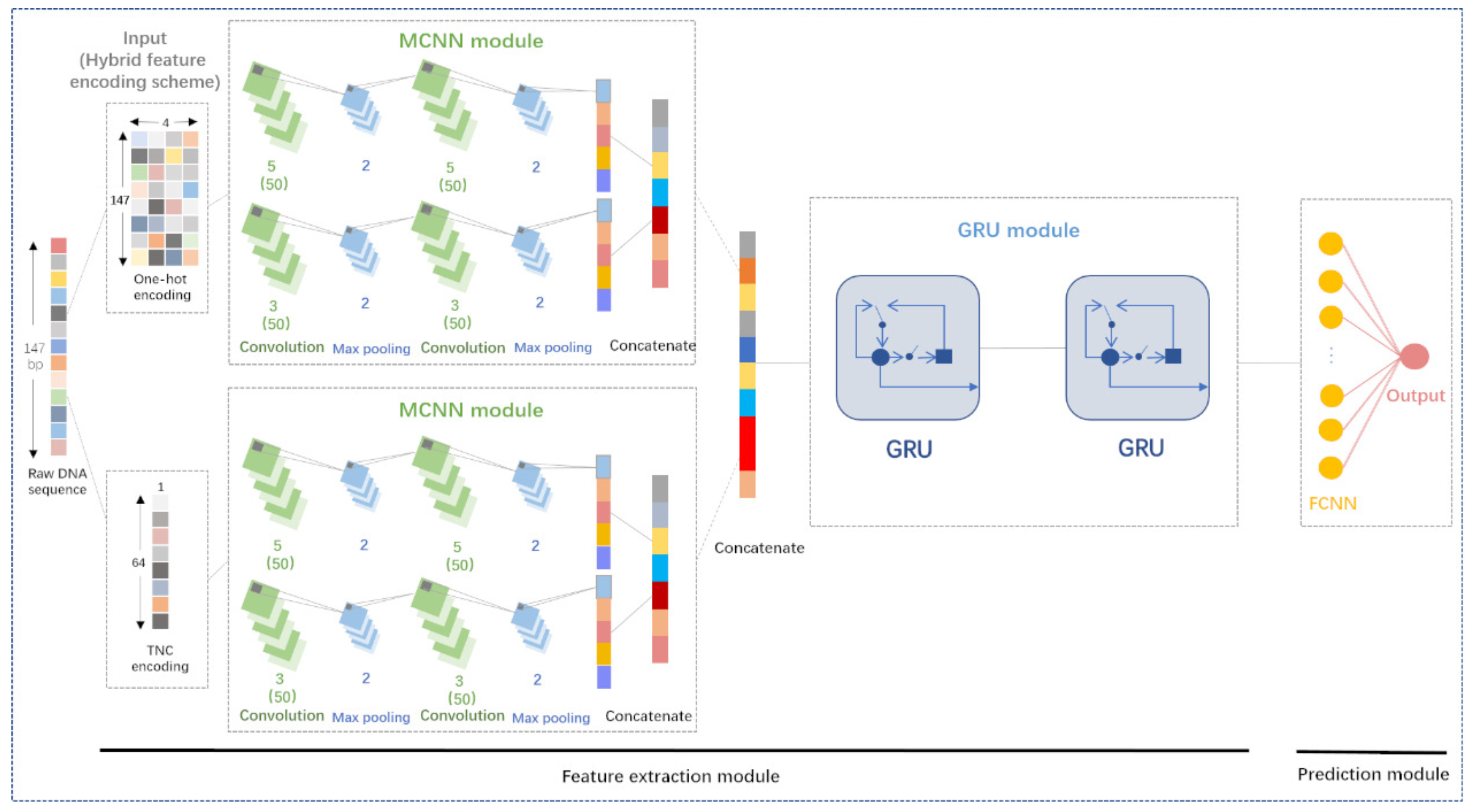
Genes | Free Full-Text | DeepNup: Prediction of Nucleosome Positioning from DNA Sequences Using Deep Neural Network

python - How to transform amino acid raw data to 3d tensor by one hot encoding by tensorflow - Stack Overflow
Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks

Conjoint Feature Representation of GO and Protein Sequence for PPI Prediction Based on an Inception RNN Attention Network: Molecular Therapy - Nucleic Acids

How Deep Learning Tools Can Help Protein Engineers Find Good Sequences | The Journal of Physical Chemistry B

A Convolutional Neural Network Using Dinucleotide One-hot Encoder for identifying DNA N6-Methyladenine Sites in the Rice Genome - ScienceDirect

DeepMHC: Deep Convolutional Neural Networks for High-performance peptide-MHC Binding Affinity Prediction | bioRxiv
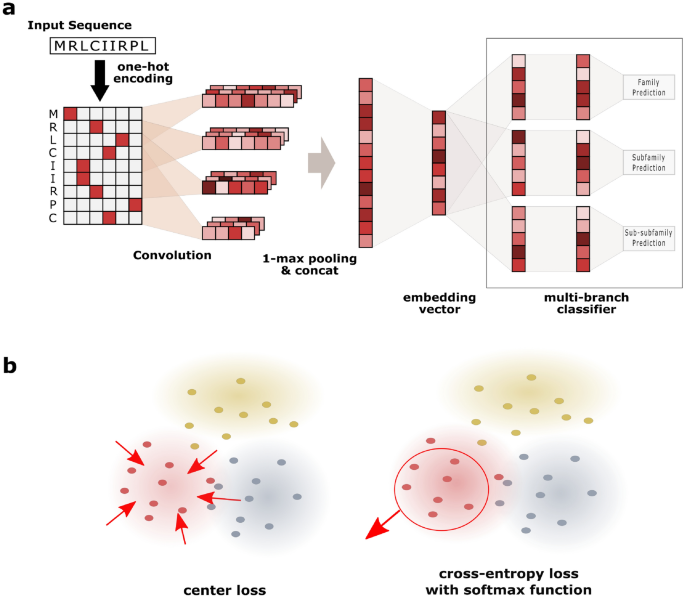
Deep hierarchical embedding for simultaneous modeling of GPCR proteins in a unified metric space | Scientific Reports
Two common encoding methods. (A) One hot encoding of base, where black... | Download Scientific Diagram

Schematic explanation of one-hot encoding, zero-padding and truncation... | Download Scientific Diagram

Structure-aware protein solubility prediction from sequence through graph convolutional network and predicted contact map | Journal of Cheminformatics | Full Text
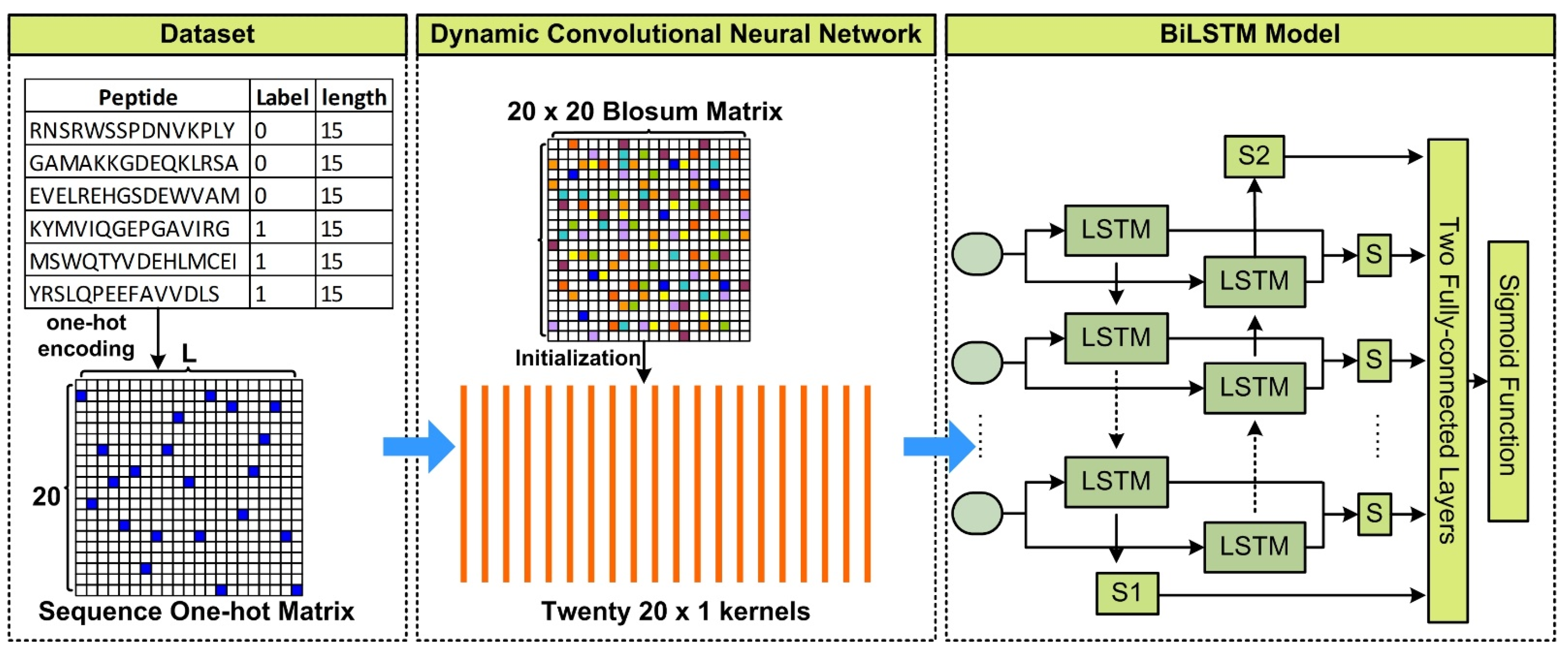
![Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ] Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/11262/1/fig-4-full.png)
Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ]