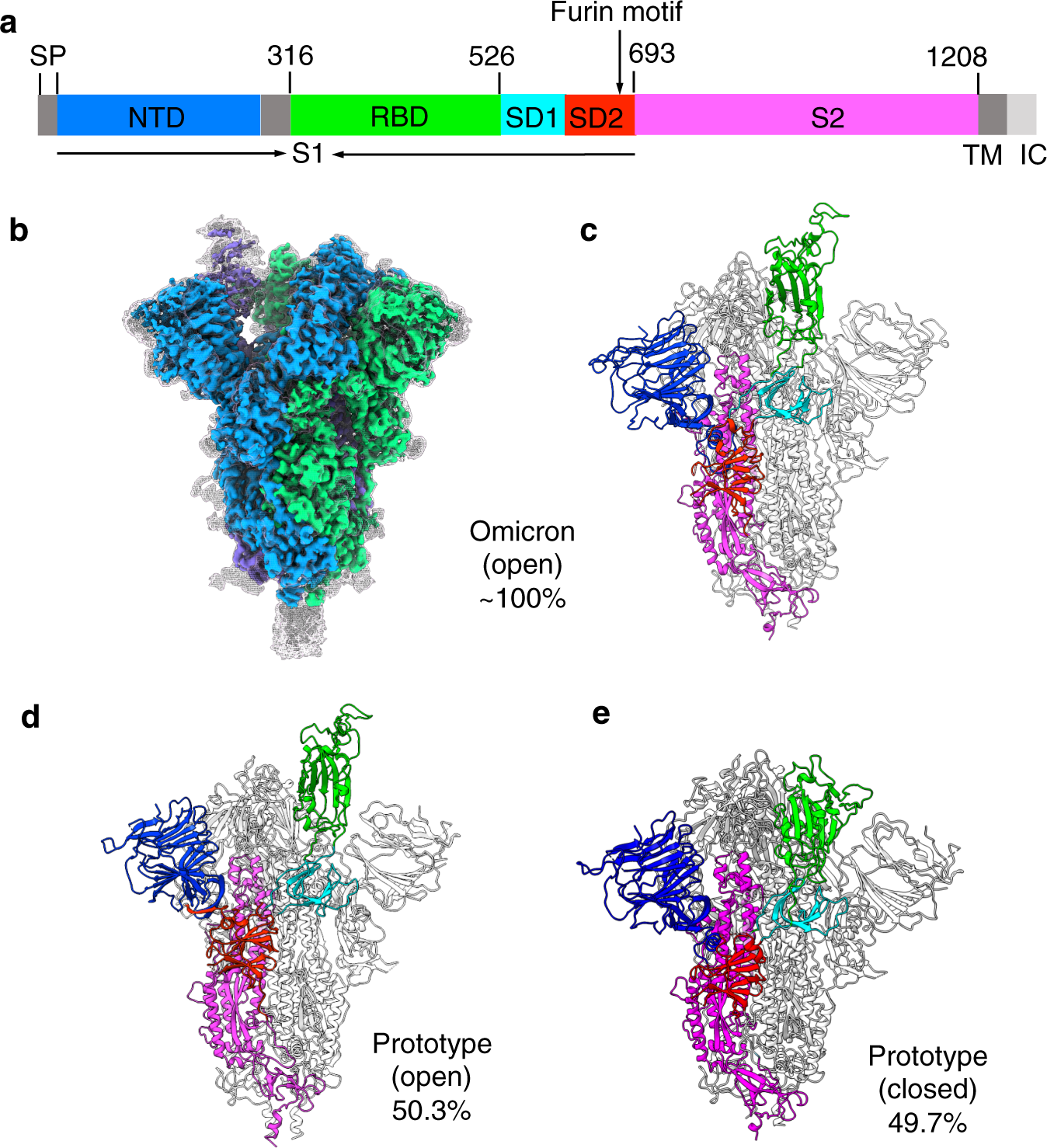
SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex | Science
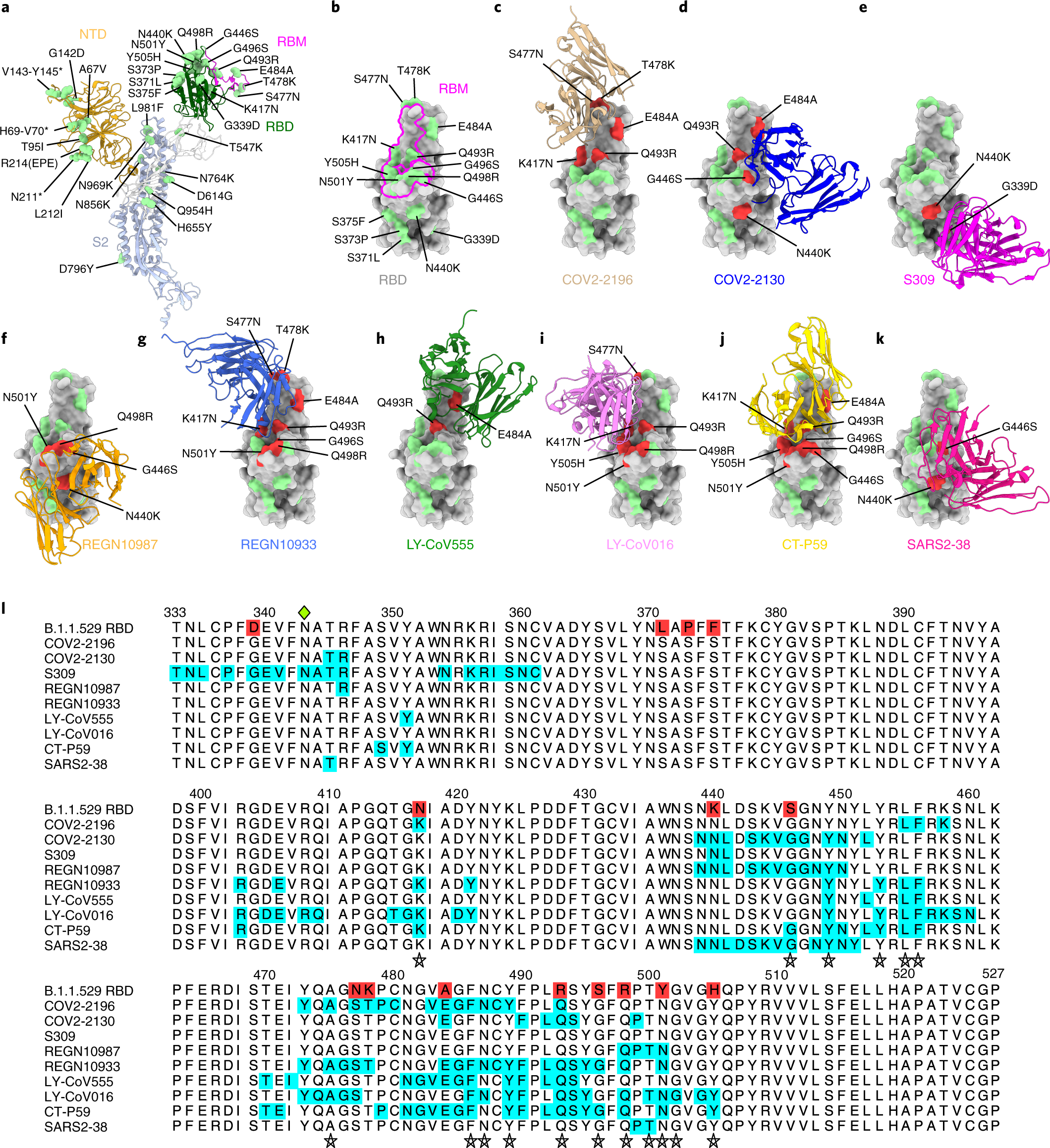
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies | Nature Medicine
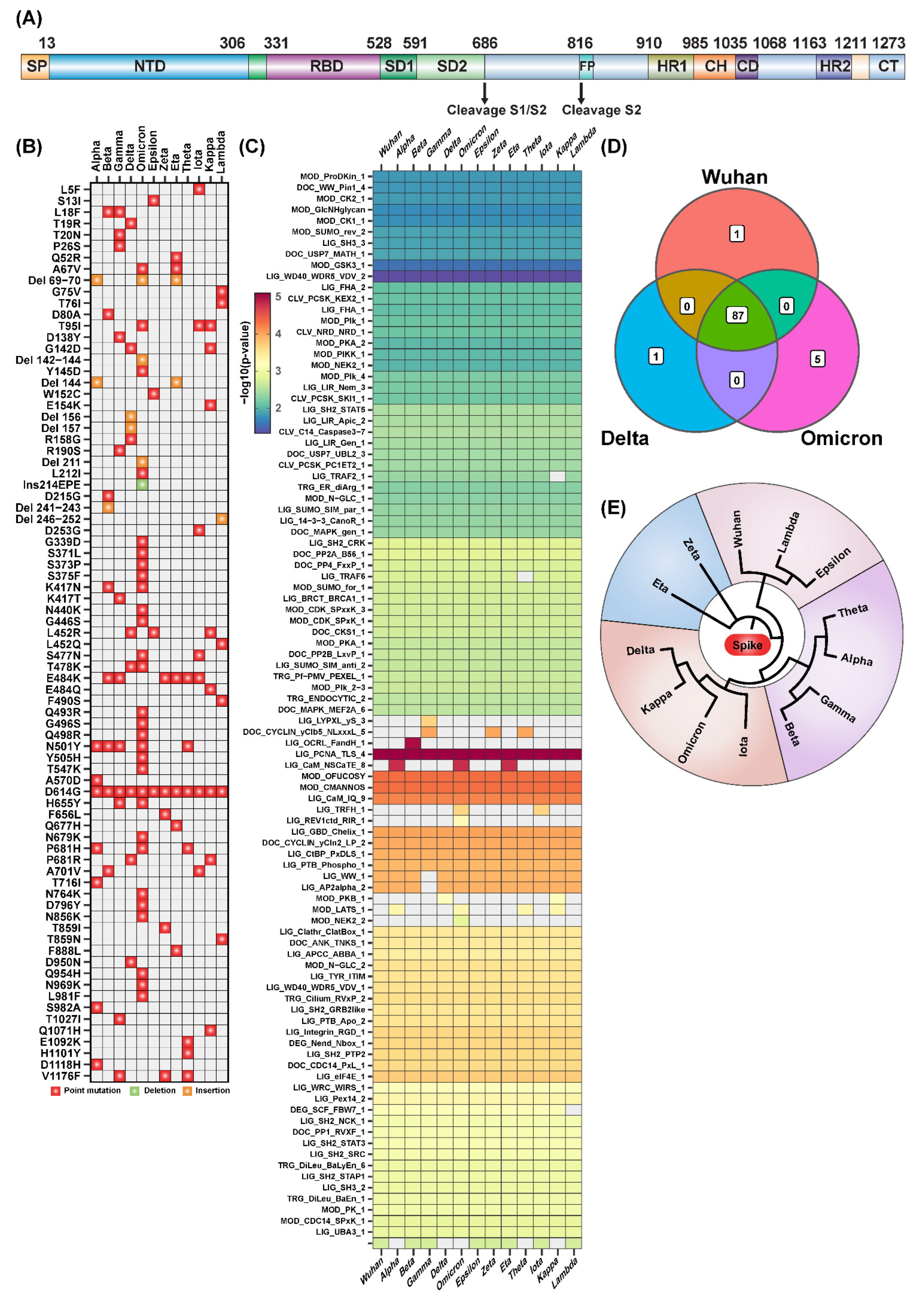
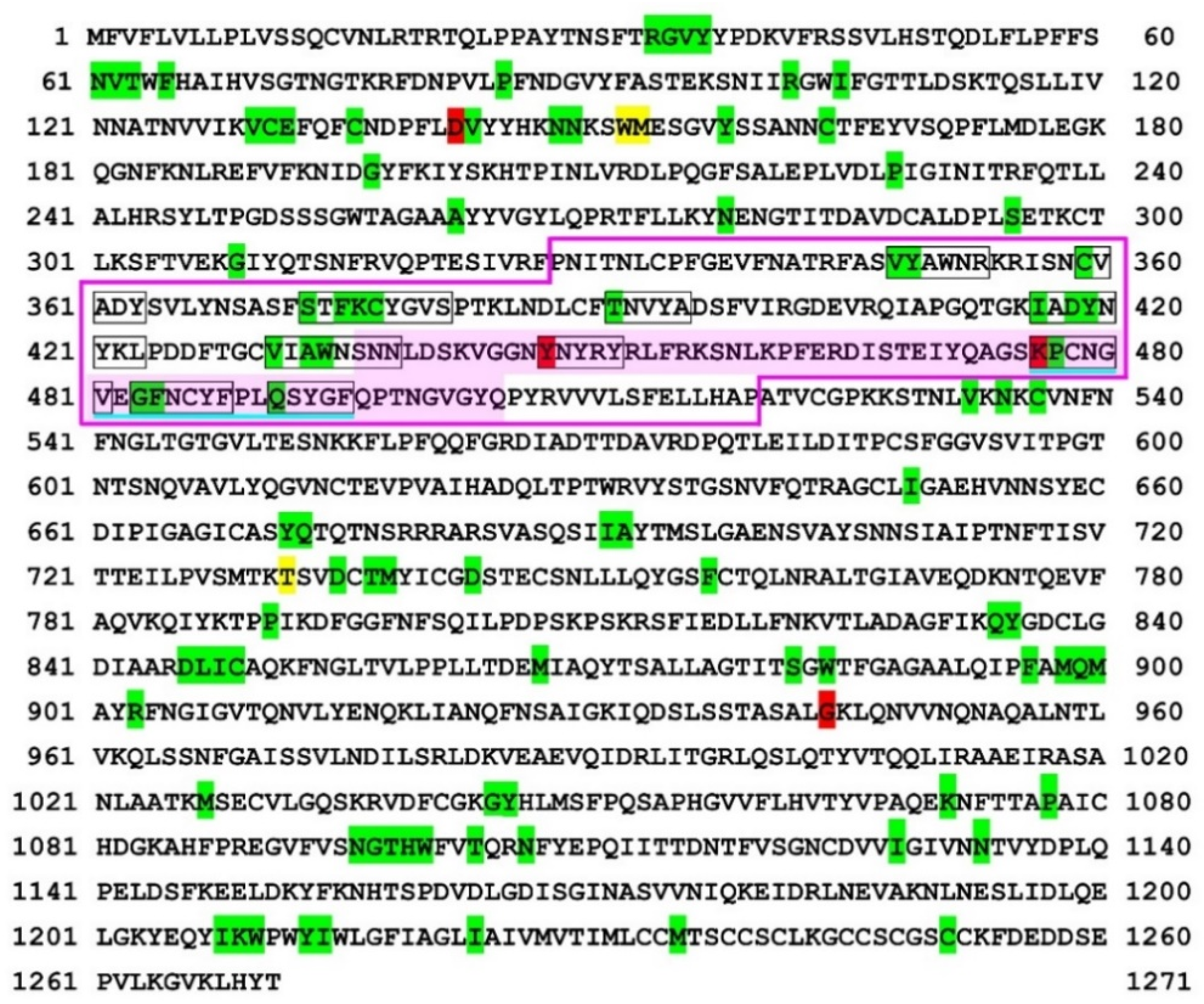
IJMS | Free Full-Text | Computational Analysis of Short Linear Motifs in the Spike Protein of SARS-CoV-2 Variants Provides Possible Clues into the Immune Hijack and Evasion Mechanisms of Omicron Variant

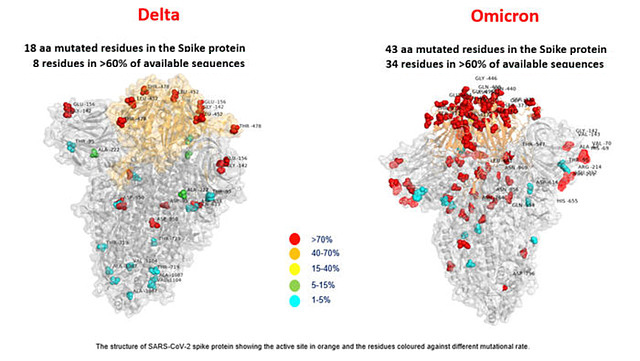
Omicron and Delta variant of SARS‐CoV‐2: A comparative computational study of spike protein - Kumar - 2022 - Journal of Medical Virology - Wiley Online Library
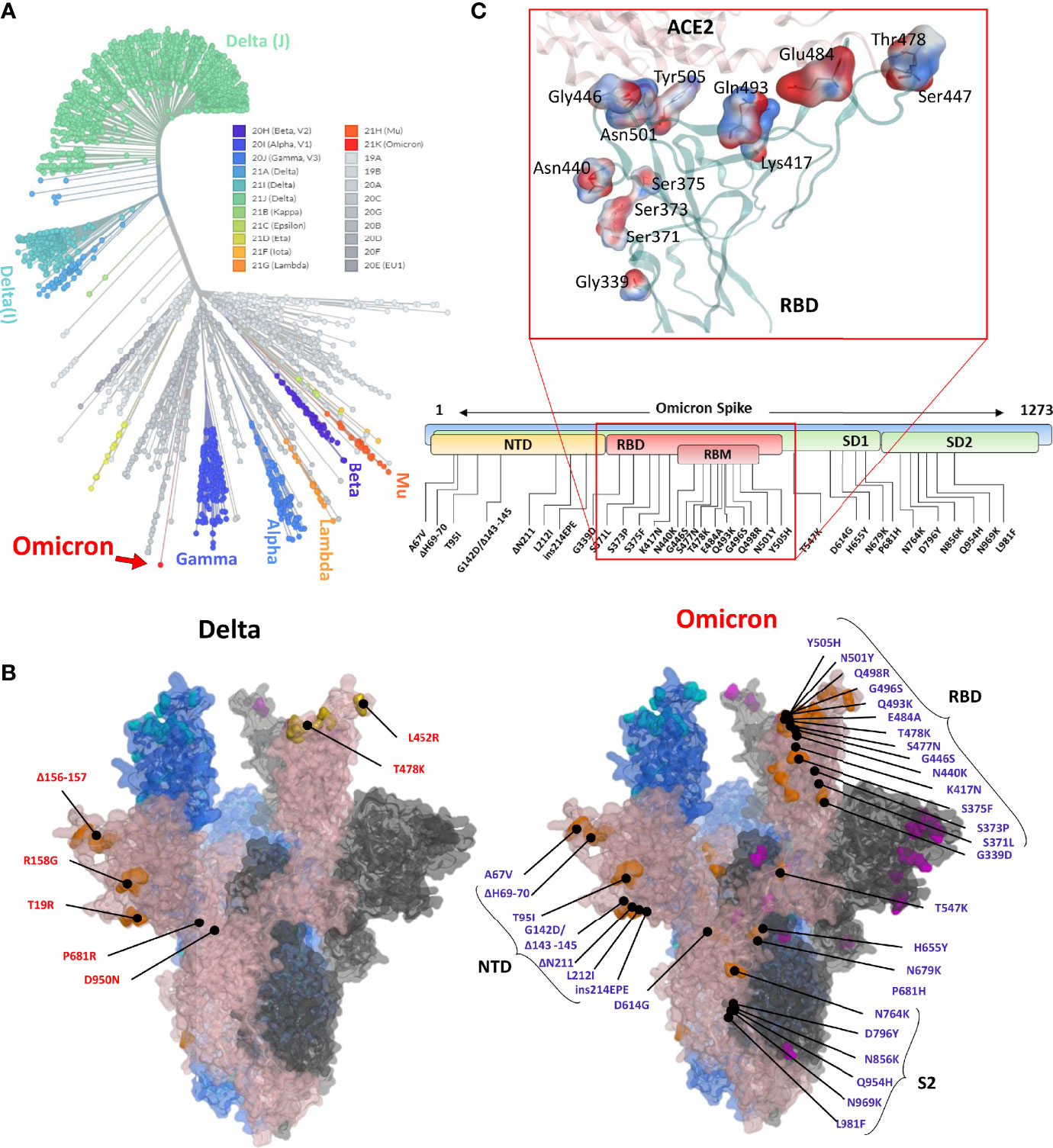
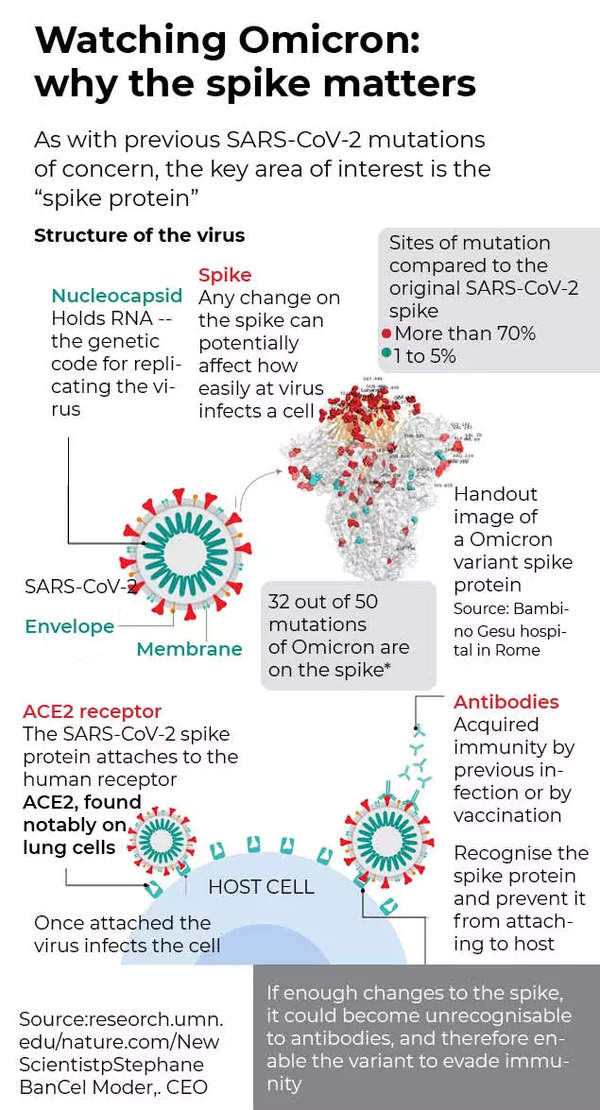
Frontiers | Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies
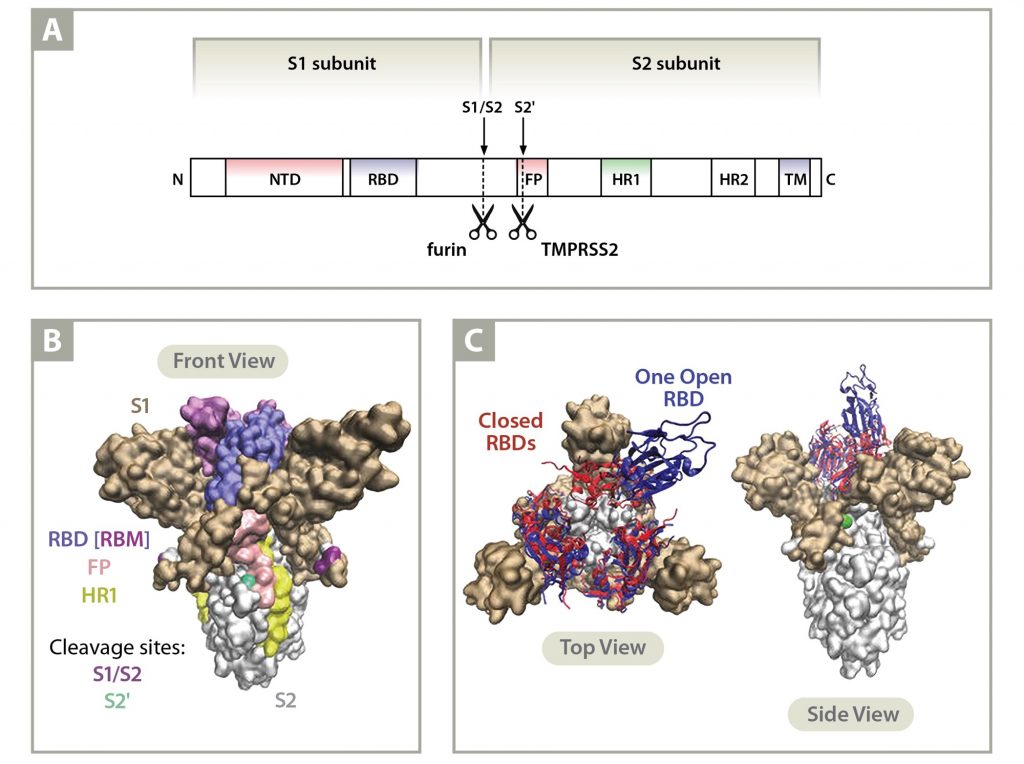
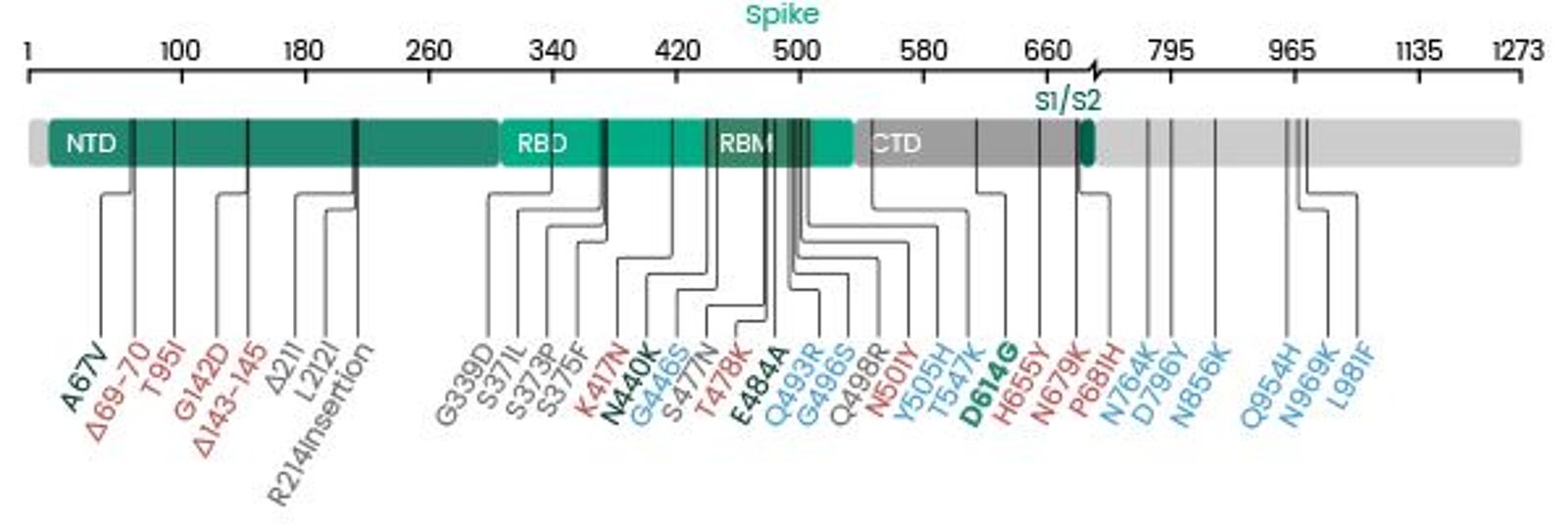
Implications of the Mutations in the Spike Protein of the Omicron Variant of Concern (VoC) of SARS-CoV-2 – Signature Science

Mutations in the spike RBD of SARS-CoV-2 omicron variant may increase infectivity without dramatically altering the efficacy of current multi-dosage vaccinations | bioRxiv

Interaction Analysis of the Spike Protein of Delta and Omicron Variants of SARS-CoV-2 with hACE2 and Eight Monoclonal Antibodies Using the Fragment Molecular Orbital Method | Journal of Chemical Information and Modeling

Determinants of Spike infectivity, processing, and neutralization in SARS-CoV-2 Omicron subvariants BA.1 and BA.2 - ScienceDirect
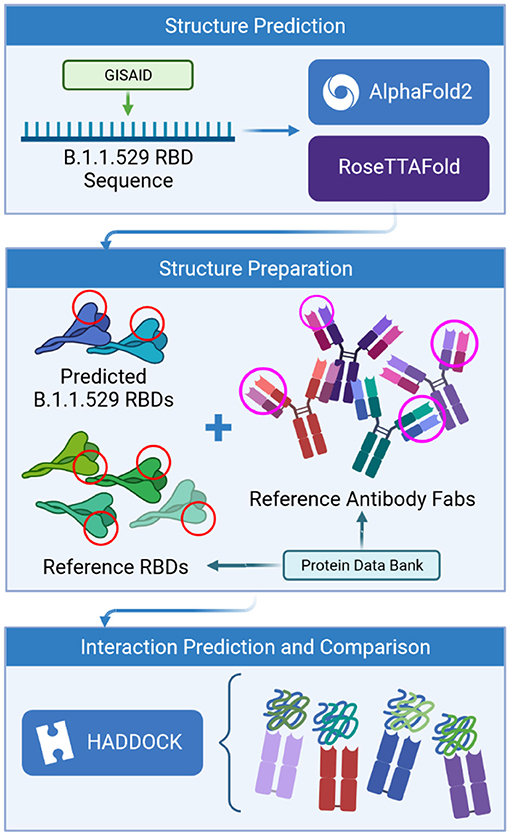
Frontiers | Predictions of the SARS-CoV-2 Omicron Variant (B.1.1.529) Spike Protein Receptor-Binding Domain Structure and Neutralizing Antibody Interactions
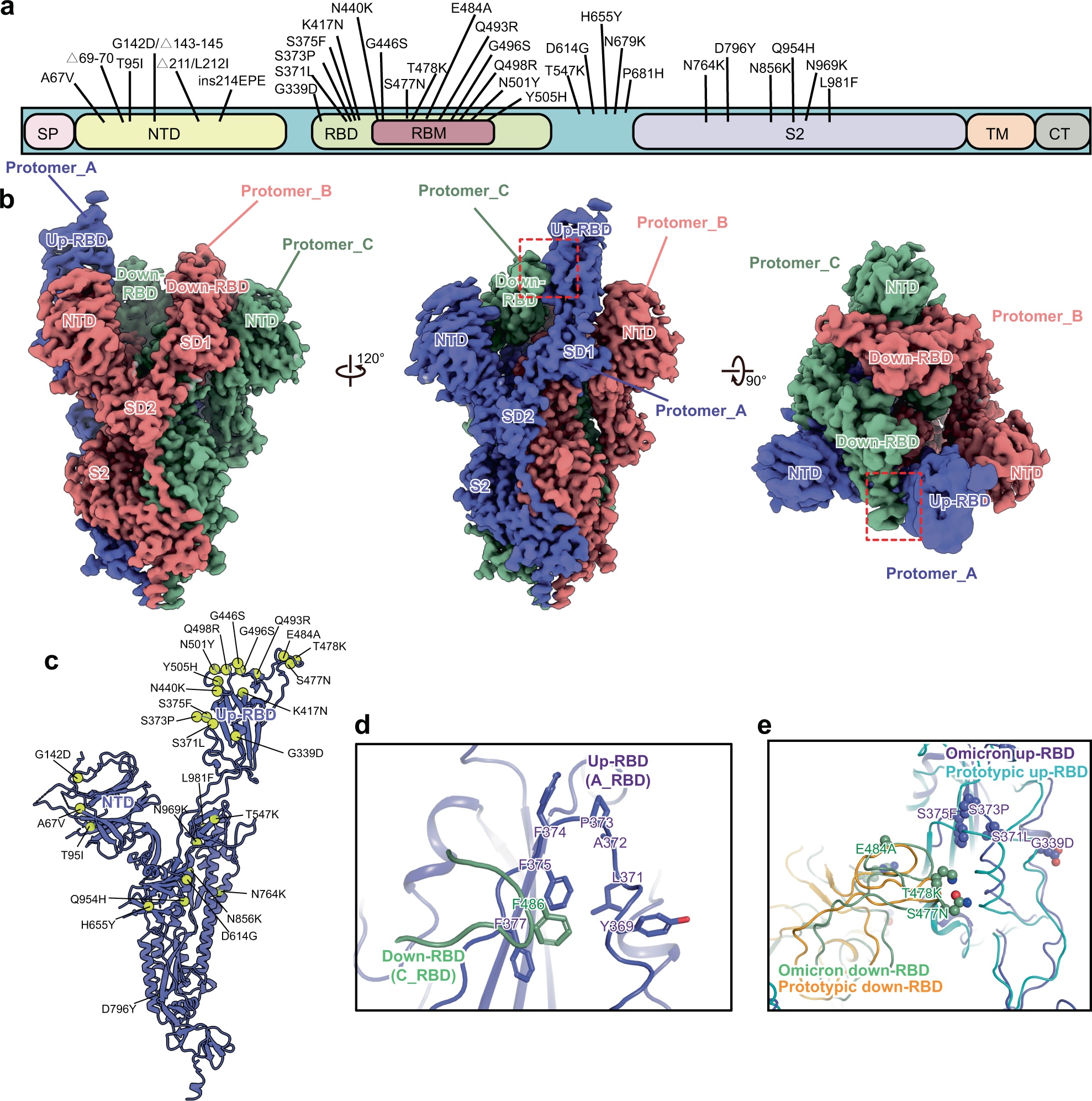
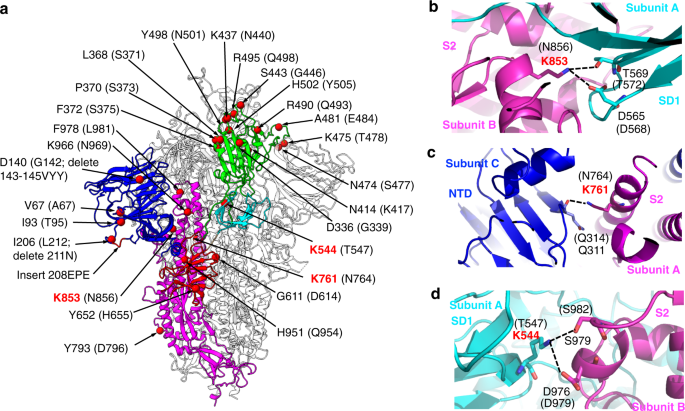
Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape | Nature Communications
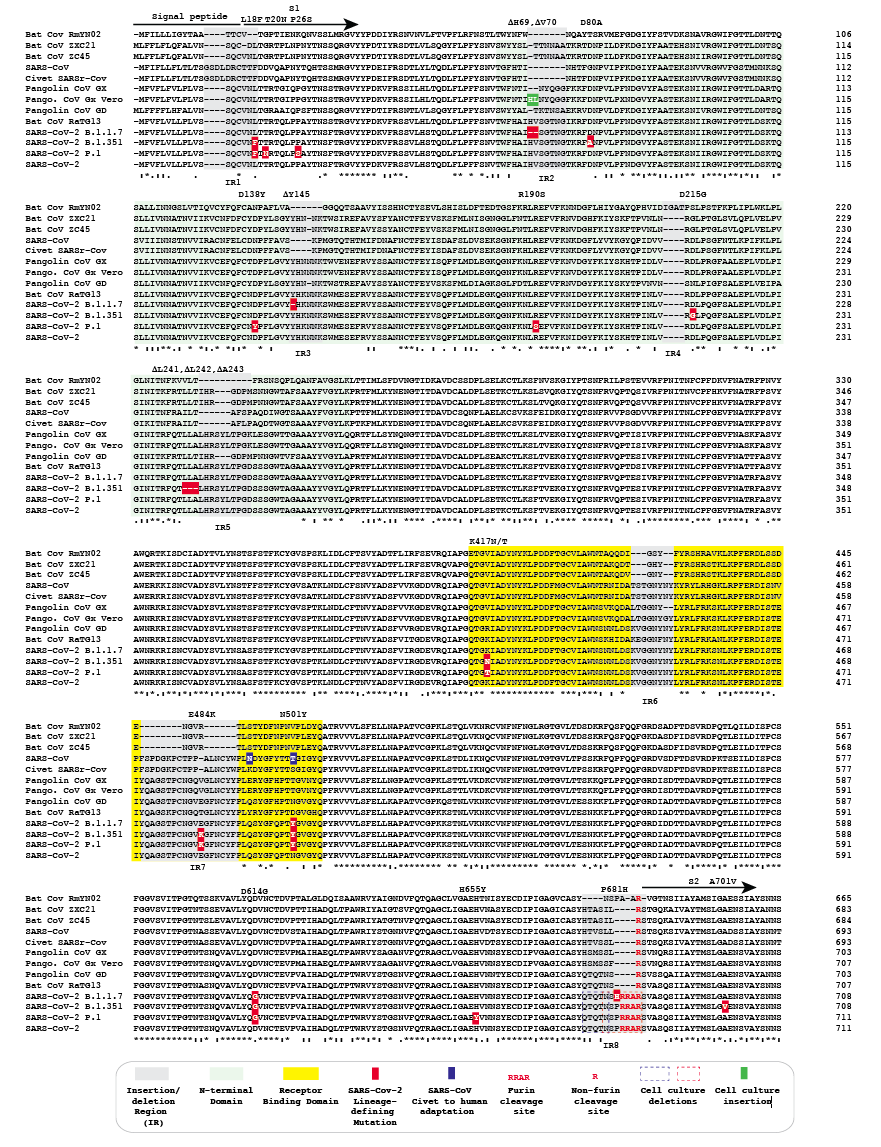
Spike protein mutations in novel SARS-CoV-2 'variants of concern' commonly occur in or near indels - Virological