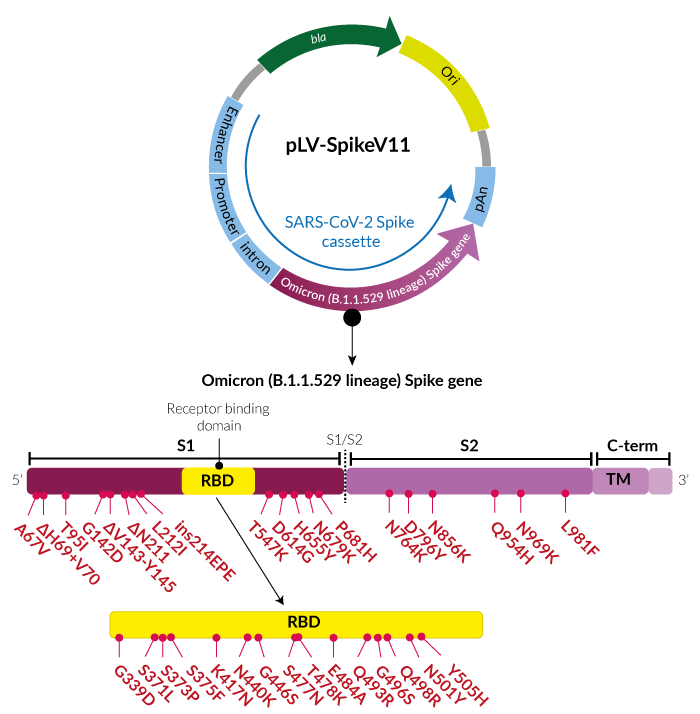
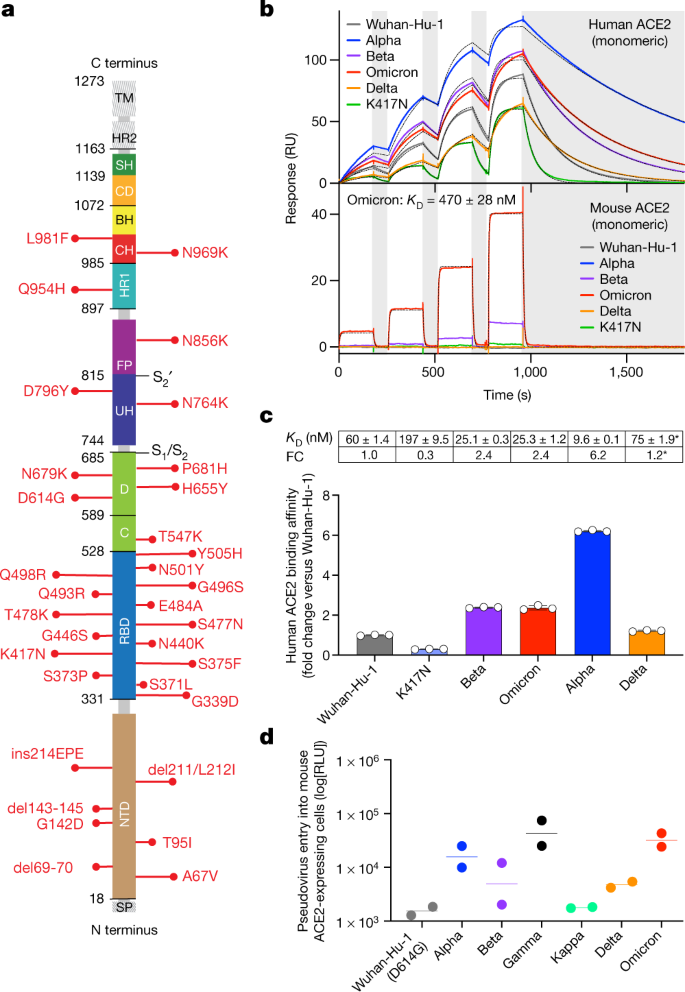
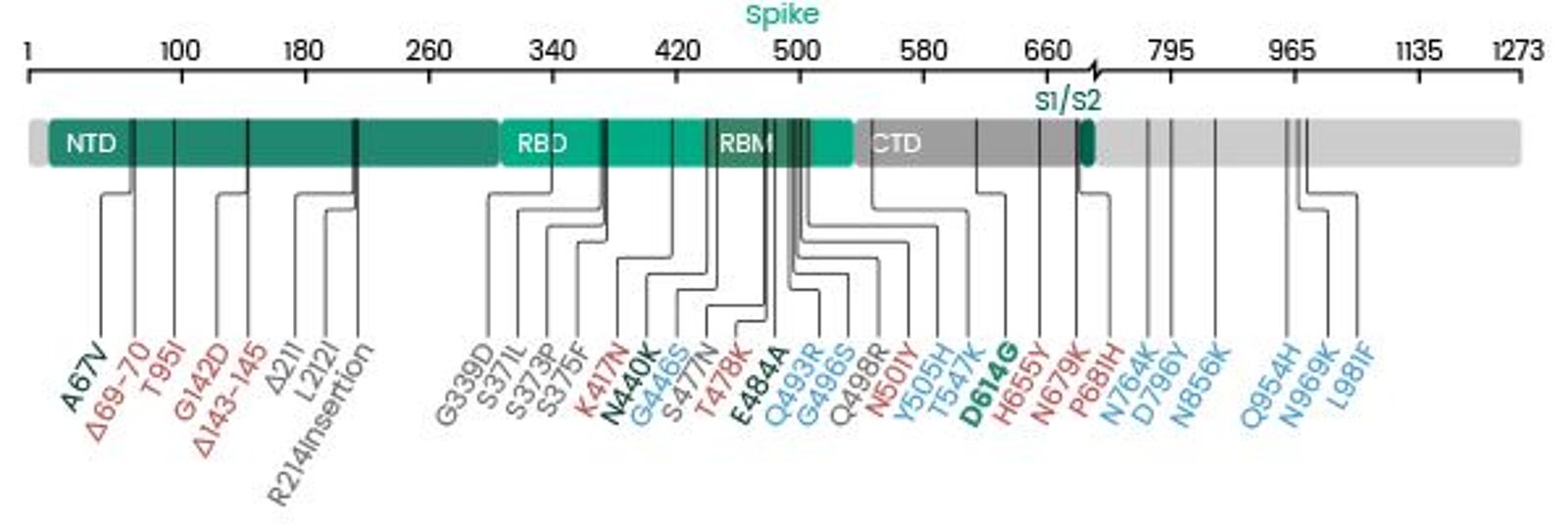
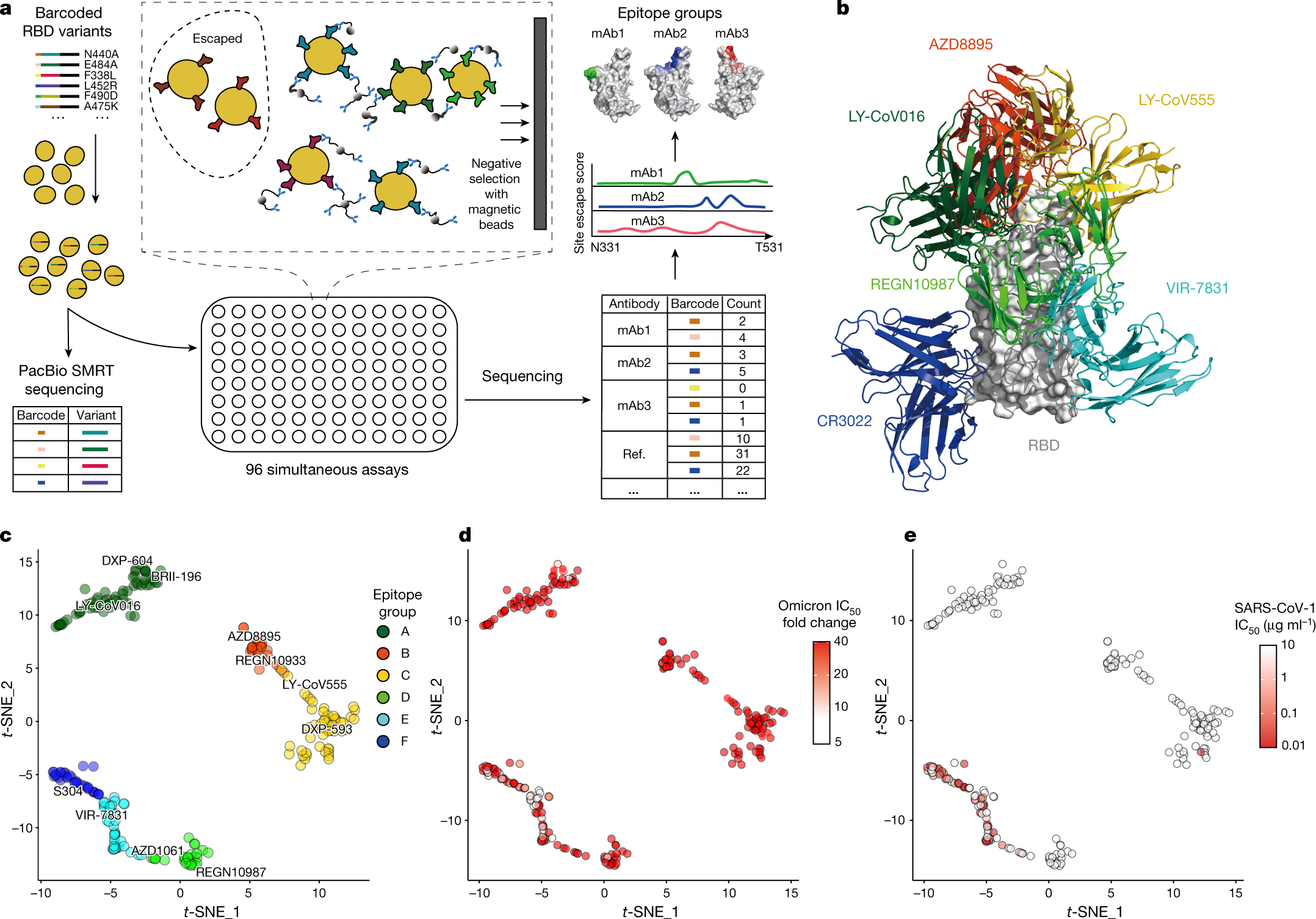
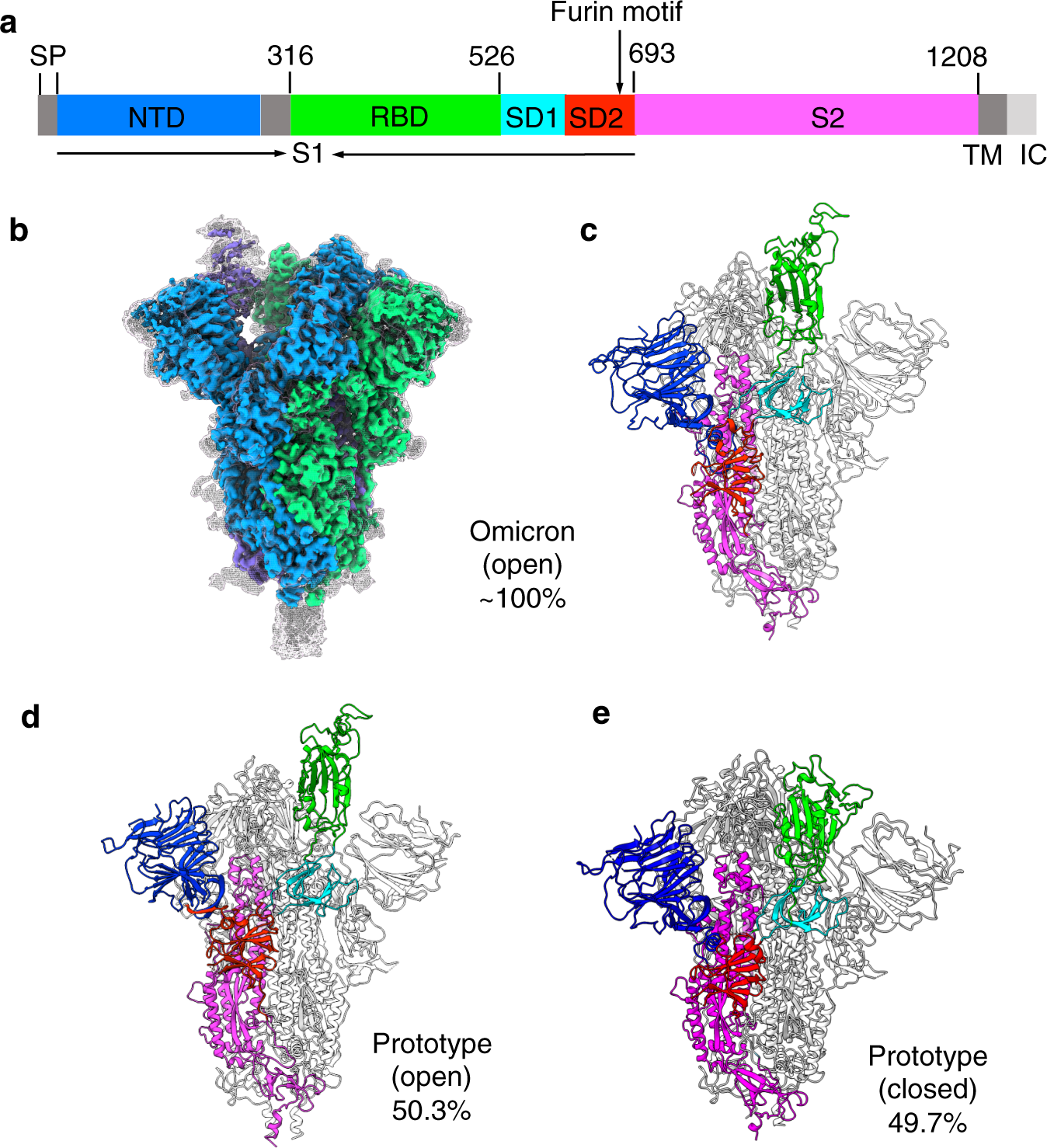
Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2 - ScienceDirect
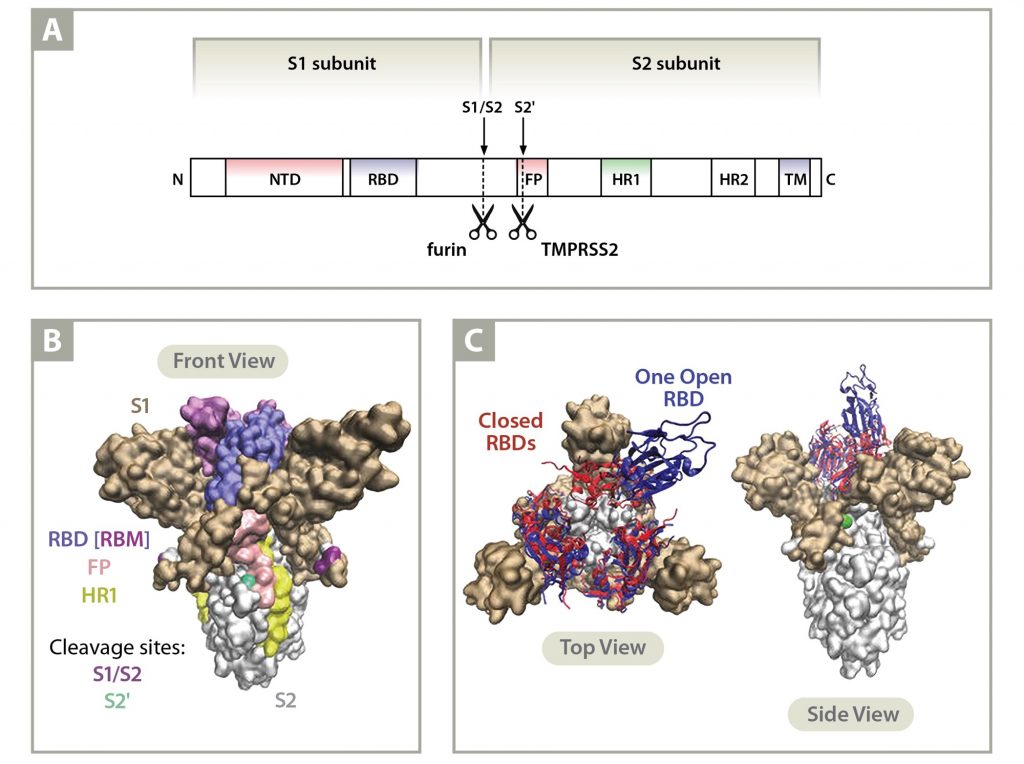
Implications of the Mutations in the Spike Protein of the Omicron Variant of Concern (VoC) of SARS-CoV-2 – Signature Science
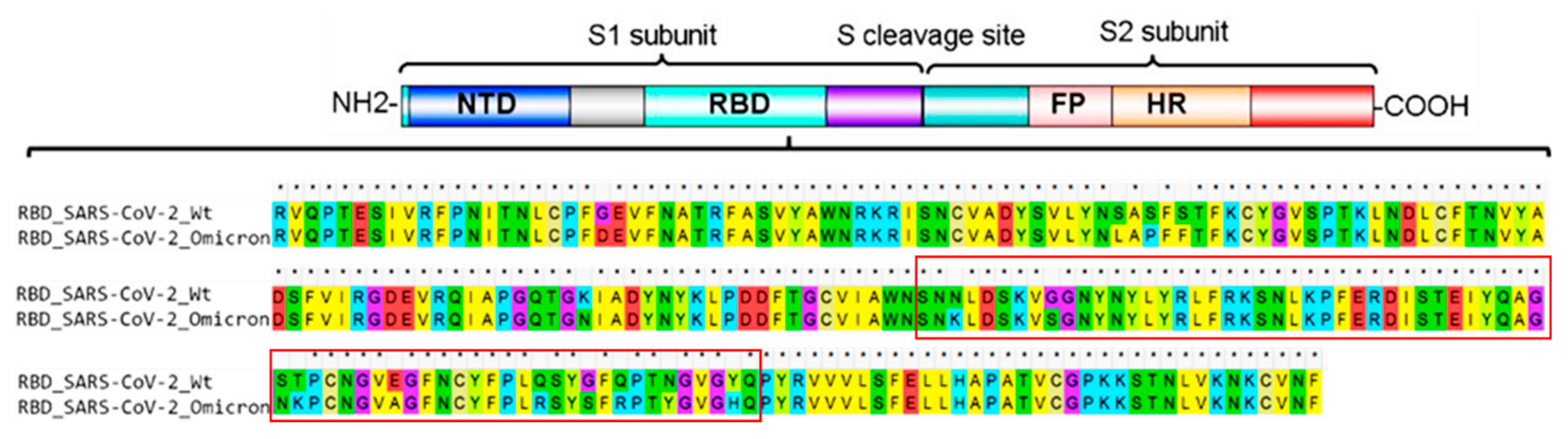
IJMS | Free Full-Text | Improved Binding Affinity of Omicron’s Spike Protein for the Human Angiotensin-Converting Enzyme 2 Receptor Is the Key behind Its Increased Virulence

Mutations in the spike RBD of SARS-CoV-2 omicron variant may increase infectivity without dramatically altering the efficacy of current multi-dosage vaccinations | bioRxiv

Analysis of a SARS-CoV-2 convalescent cohort identified a common strategy for escape of vaccine-induced anti-RBD antibodies by Beta and Omicron variants - eBioMedicine
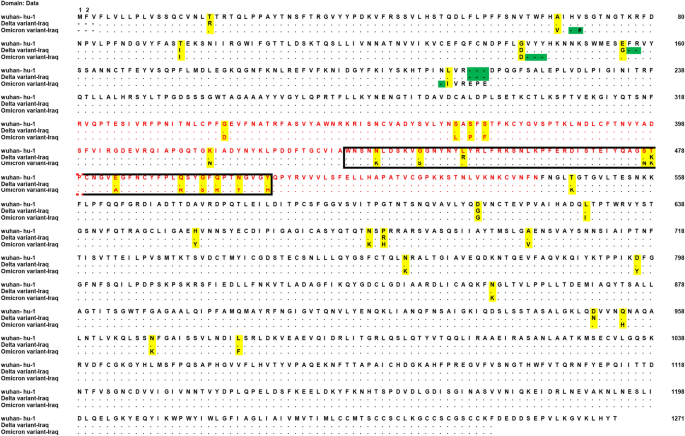
Molecular and computational analysis of spike protein of newly emerged omicron variant in comparison to the delta variant of SARS-CoV-2 in Iraq | SpringerLink
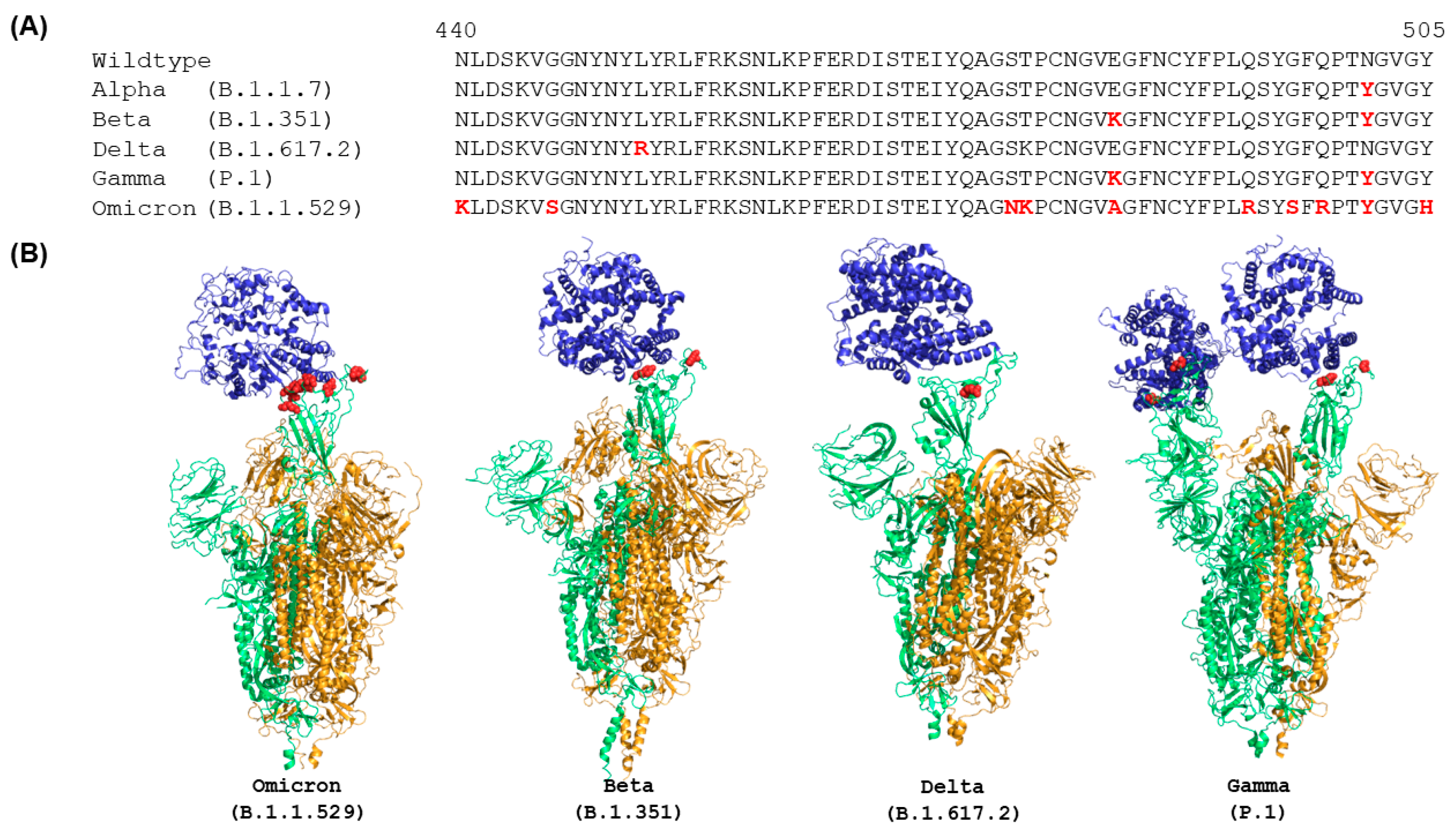
IJMS | Free Full-Text | Distinct Conformations of SARS-CoV-2 Omicron Spike Protein and Its Interaction with ACE2 and Antibody

Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles | PNAS

Interaction Analysis of the Spike Protein of Delta and Omicron Variants of SARS-CoV-2 with hACE2 and Eight Monoclonal Antibodies Using the Fragment Molecular Orbital Method | Journal of Chemical Information and Modeling

Antigenicity comparison of SARS‐CoV‐2 Omicron sublineages with other variants contained multiple mutations in RBD - Li - 2022 - MedComm - Wiley Online Library
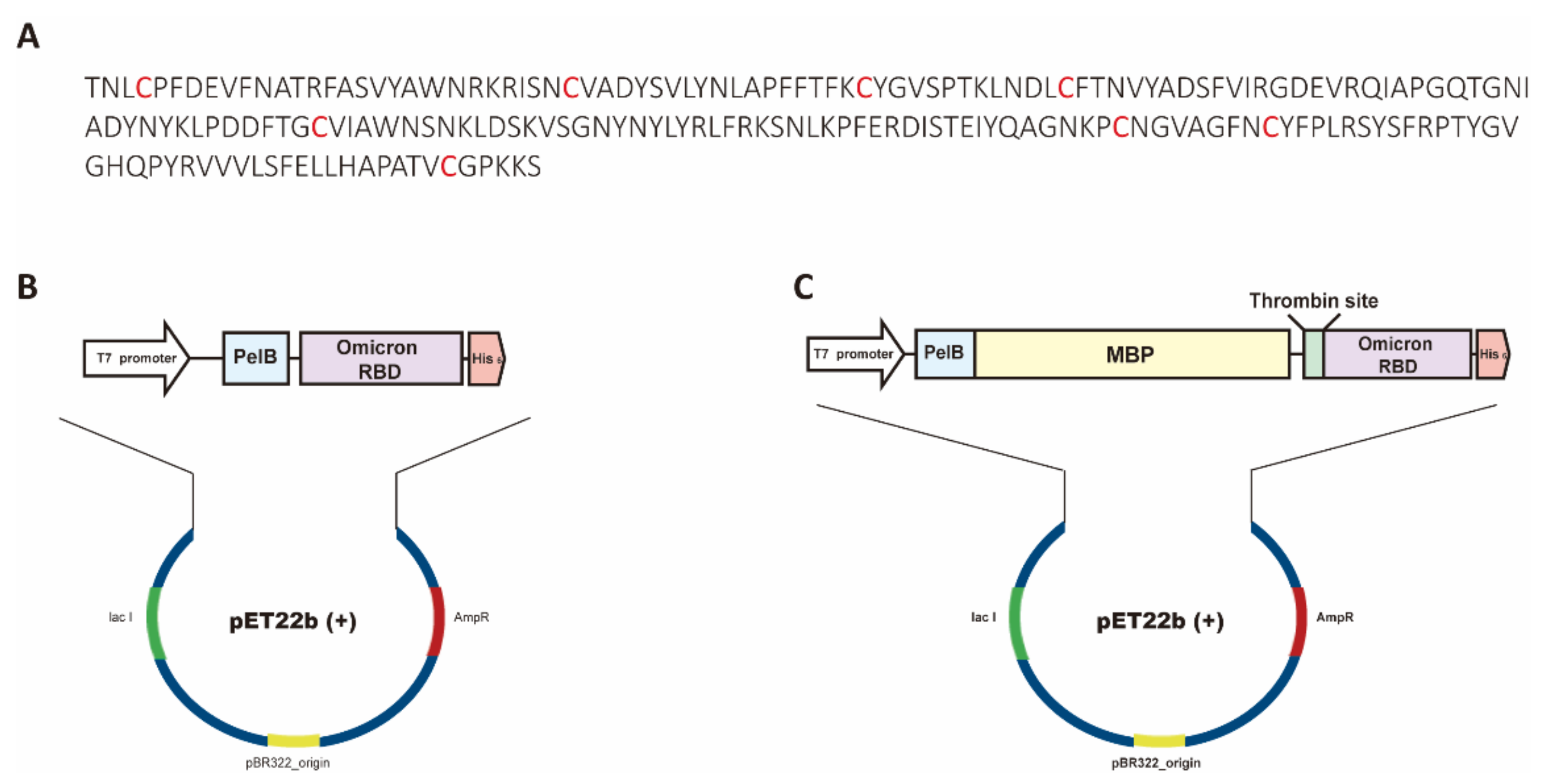
Bioengineering | Free Full-Text | Functional Expression of the Recombinant Spike Receptor Binding Domain of SARS-CoV-2 Omicron in the Periplasm of Escherichia coli
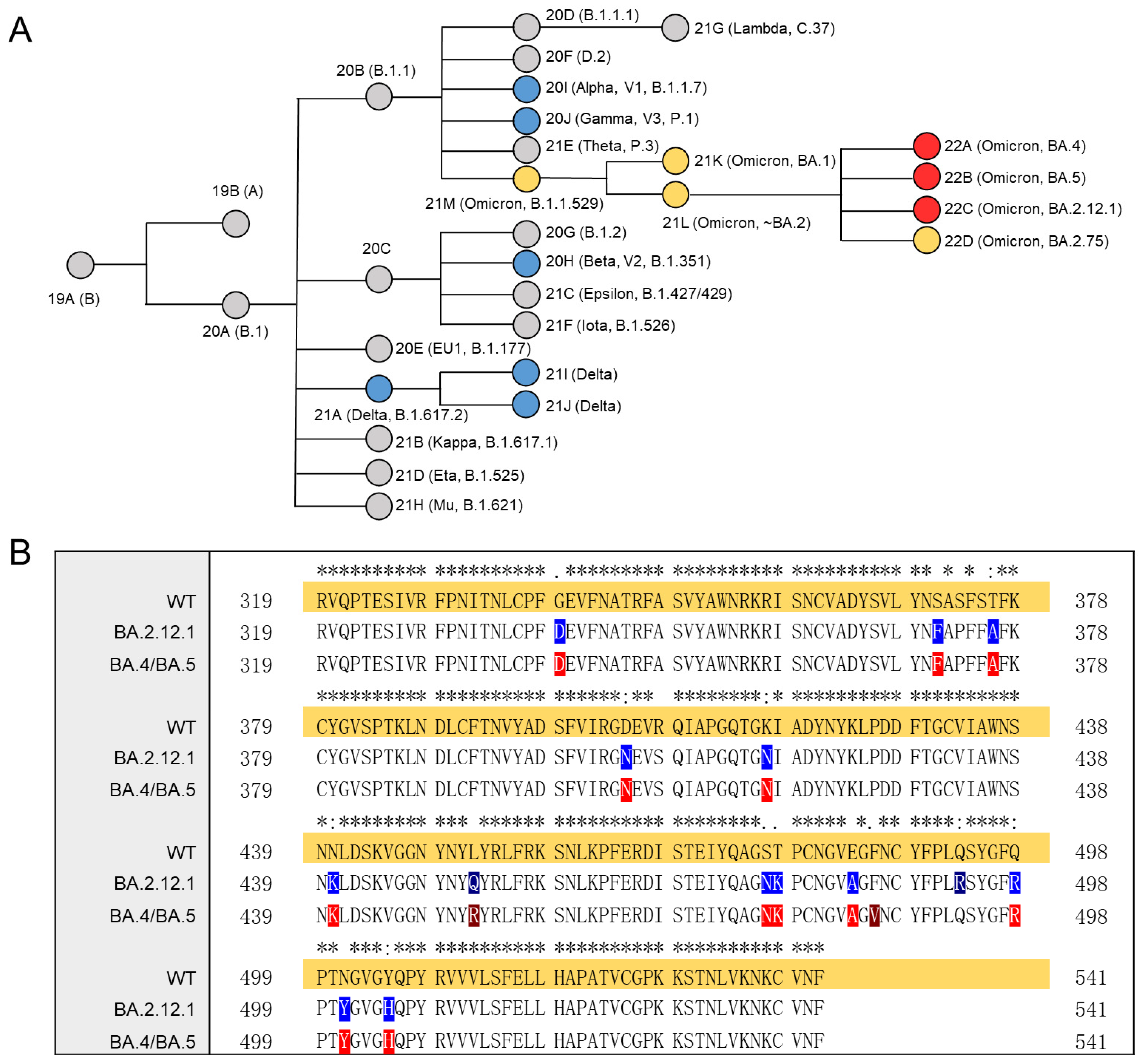
Viruses | Free Full-Text | Structural Characteristics of Heparin Binding to SARS-CoV-2 Spike Protein RBD of Omicron Sub-Lineages BA.2.12.1, BA.4 and BA.5

Sequence analysis of the emerging SARS‐CoV‐2 variant Omicron in South Africa - Wang - 2022 - Journal of Medical Virology - Wiley Online Library

Characterization of SARS-CoV-2 Omicron spike RBD reveals significantly decreased stability, severe evasion of neutralizing-antibody recognition but unaffected engagement by decoy ACE2 modified for enhanced RBD binding | Signal Transduction and Targeted ...
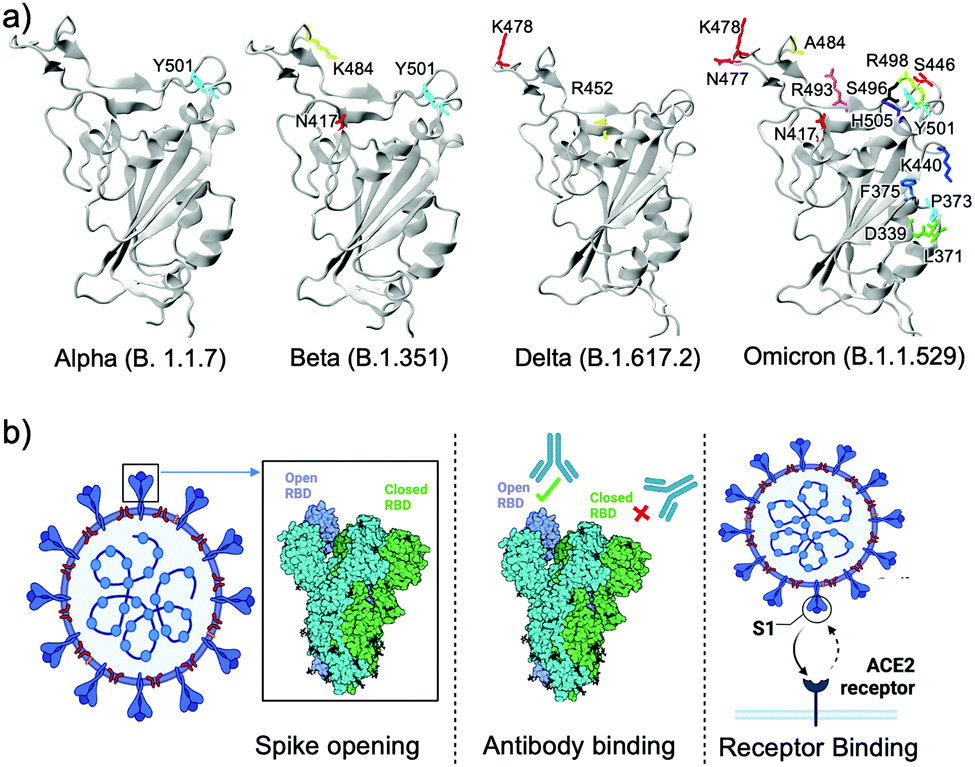
Significance of the RBD mutations in the SARS-CoV-2 omicron: from spike opening to antibody escape and cell attachment - Physical Chemistry Chemical Physics (RSC Publishing) DOI:10.1039/D2CP00169A
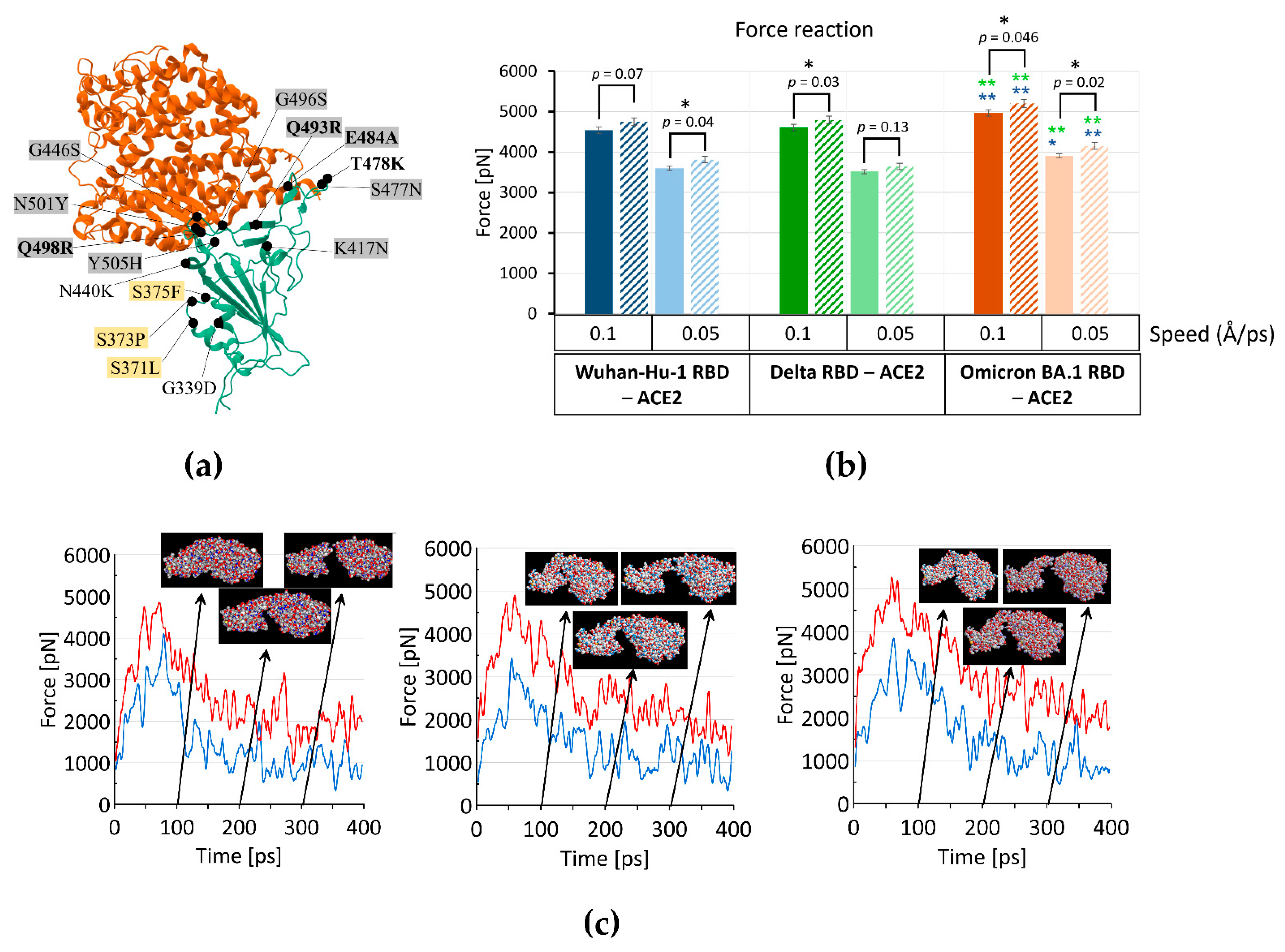
IJMS | Free Full-Text | The Increased Amyloidogenicity of Spike RBD and pH-Dependent Binding to ACE2 May Contribute to the Transmissibility and Pathogenic Properties of SARS-CoV-2 Omicron as Suggested by In Silico
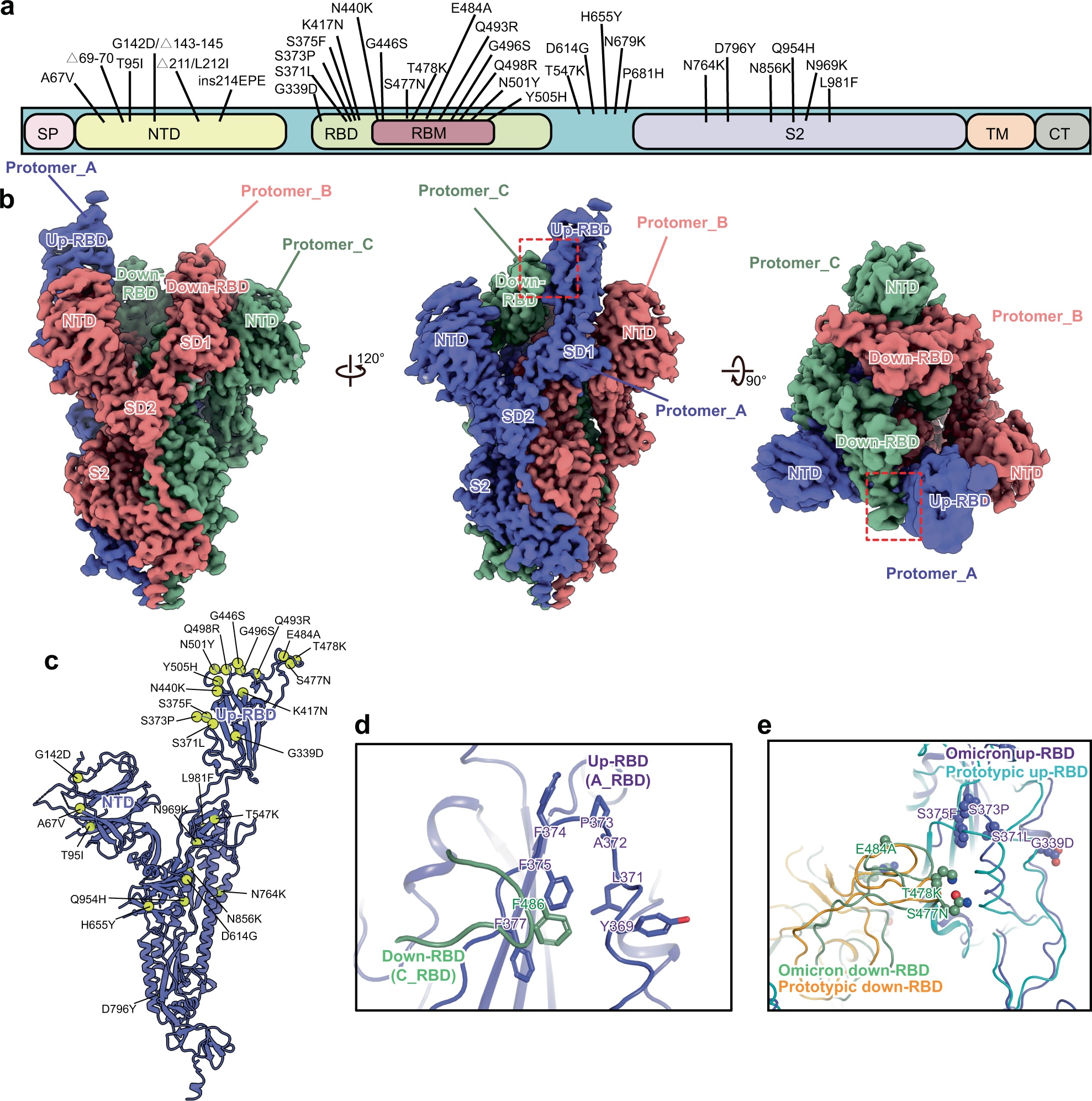
Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape | Nature Communications