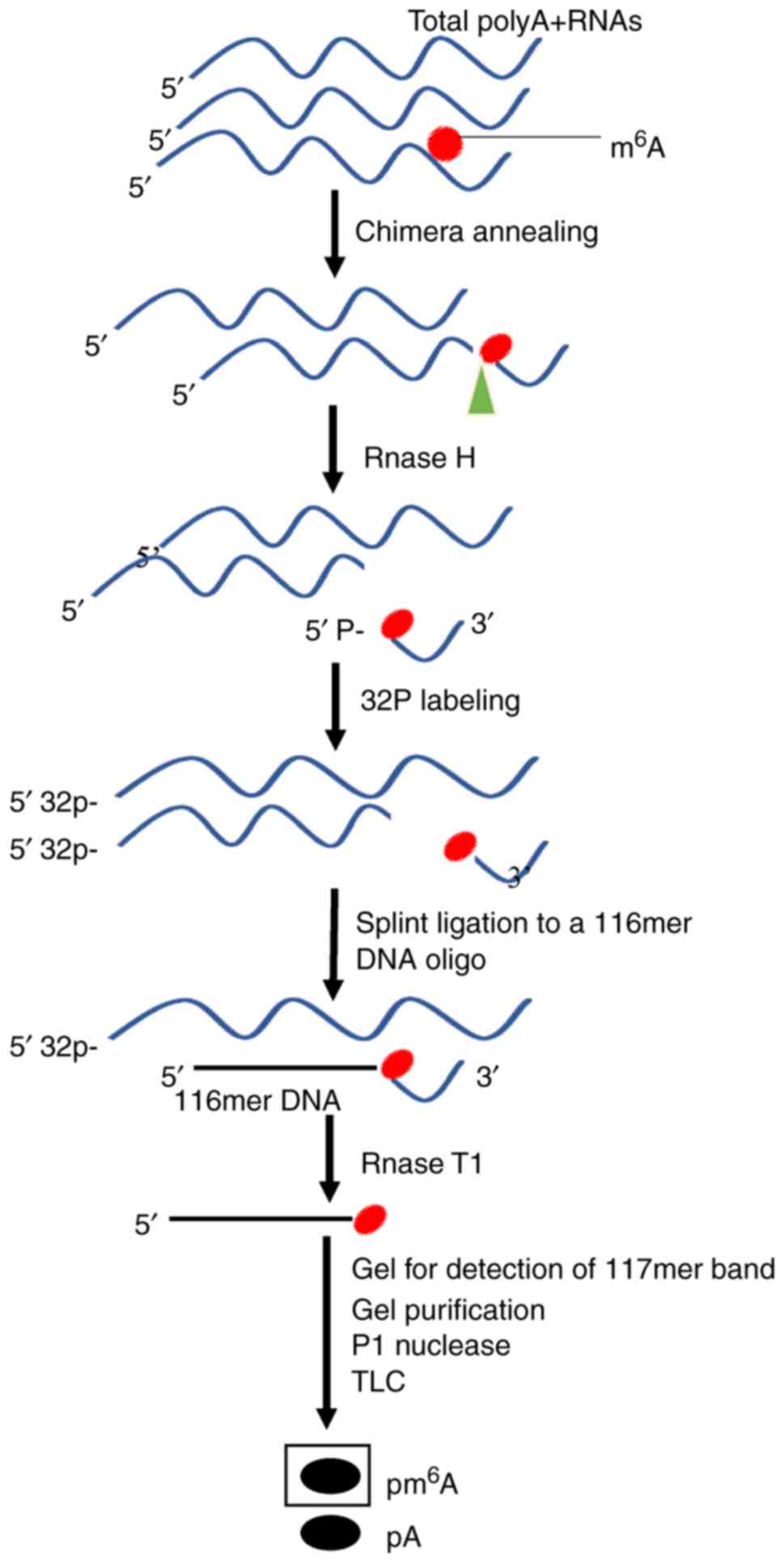
Development and validation of m6A-LAIC-seq protocol. (a) Schematic of... | Download Scientific Diagram

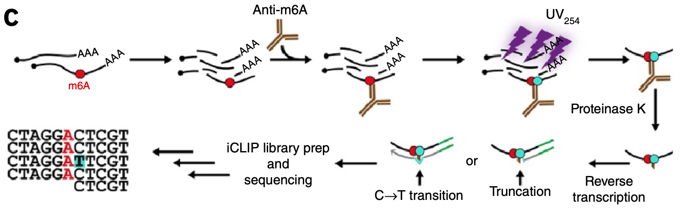
Conserved reduction of m6A RNA modifications during aging and neurodegeneration is linked to changes in synaptic transcripts | PNAS
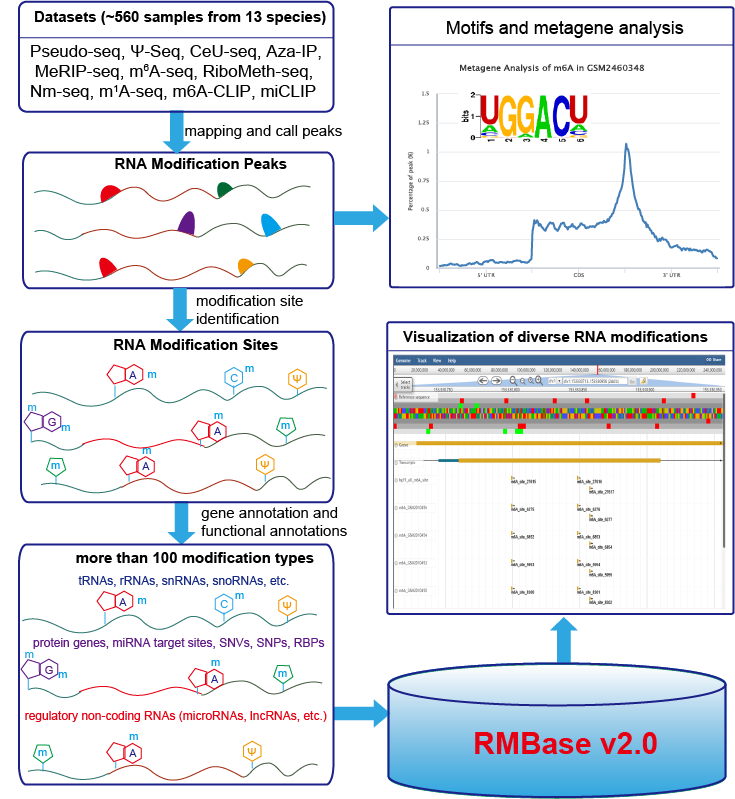
RNA Modification Database, RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data (Pseudo-seq, Ψ-seq, CeU- seq, Aza-IP, MeRIP-seq, m6A-seq, RiboMeth-seq, m1A-seq).
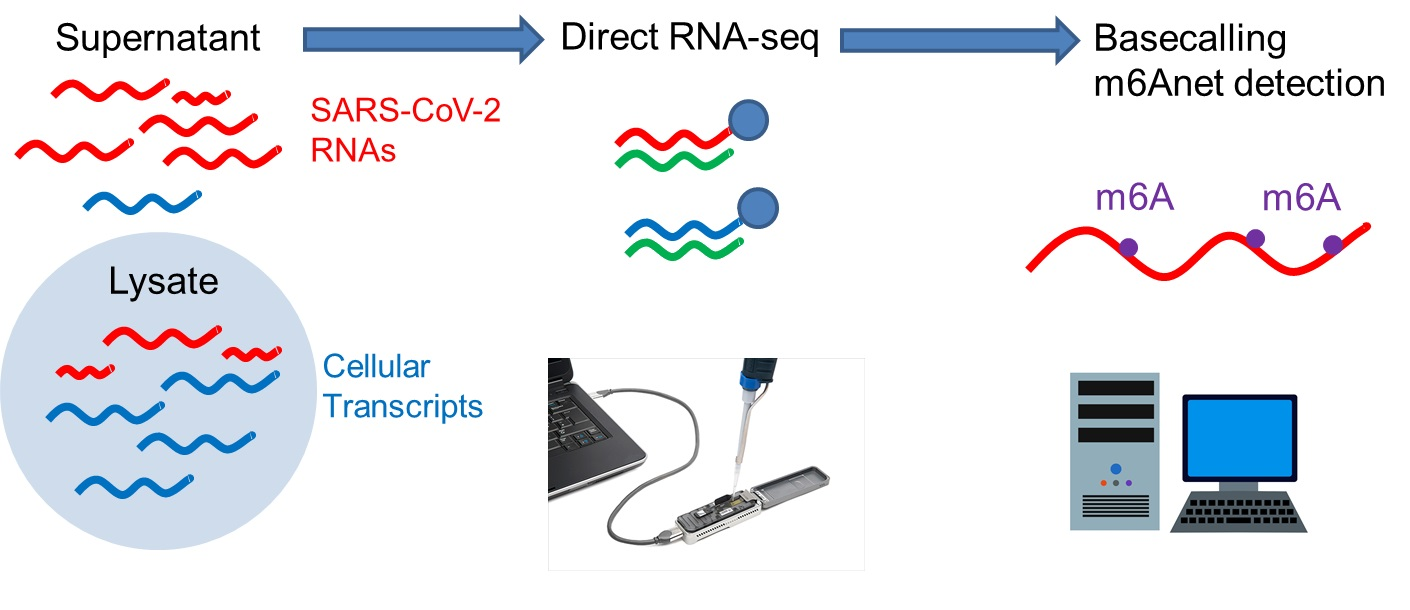
Viruses | Free Full-Text | Direct RNA Sequencing Reveals SARS-CoV-2 m6A Sites and Possible Differential DRACH Motif Methylation among Variants
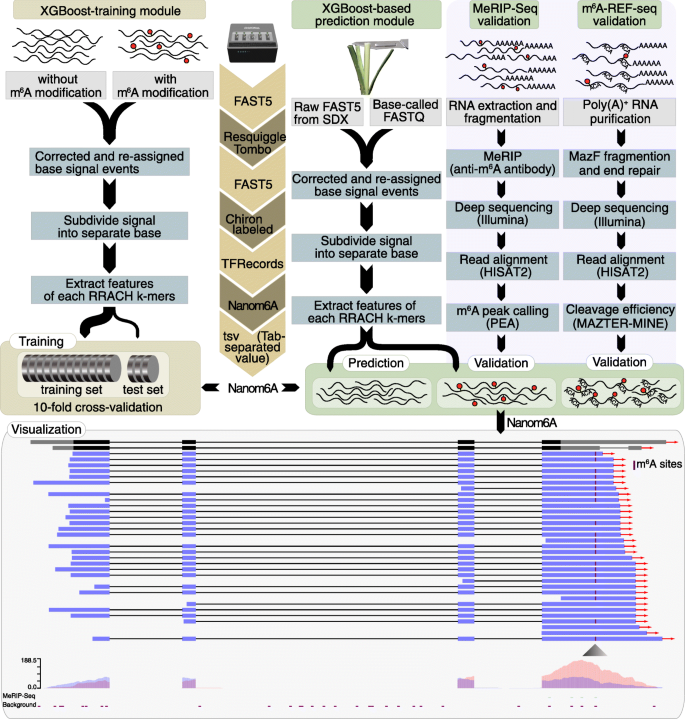
Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing | Genome Biology | Full Text

Interactions between RNA m6A modification, alternative splicing, and poly(A) tail revealed by MePAIso-seq2 | bioRxiv

Transcriptome-wide approaches to map m6A and m5C in RNA. (a) MeRIP-seq... | Download Scientific Diagram

Relevance of N6-methyladenosine regulators for transcriptome: Implications for development and the cardiovascular system - Journal of Molecular and Cellular Cardiology

scDART-seq reveals distinct m6A signatures and mRNA methylation heterogeneity in single cells - ScienceDirect

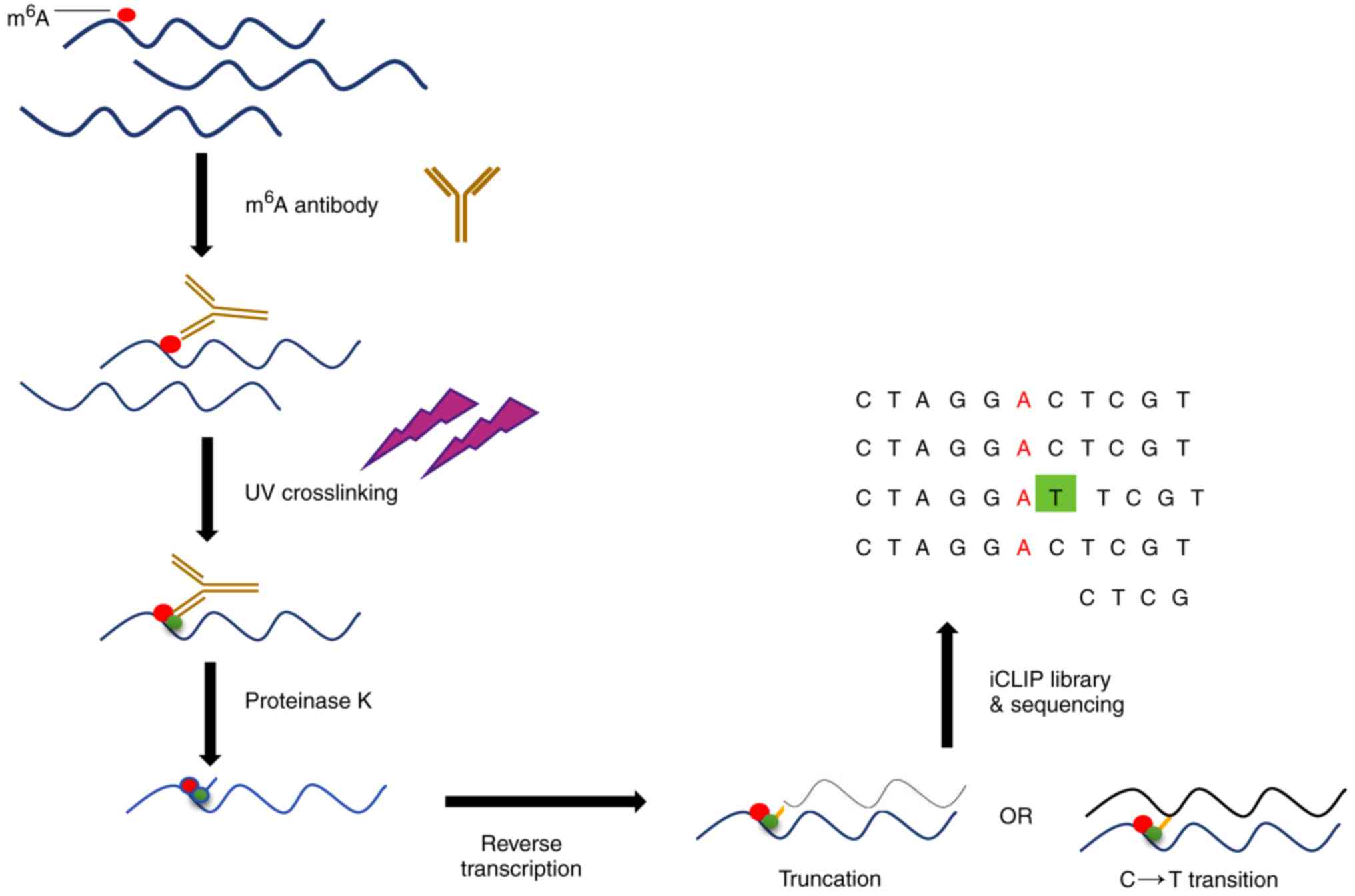
High‐Resolution N6‐Methyladenosine (m6A) Map Using Photo‐Crosslinking‐Assisted m6A Sequencing - Chen - 2015 - Angewandte Chemie International Edition - Wiley Online Library

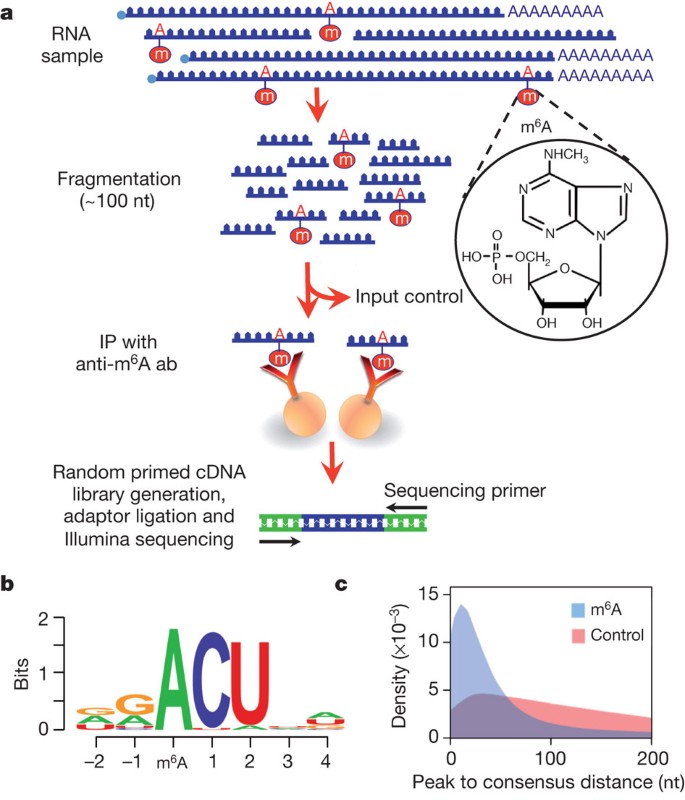
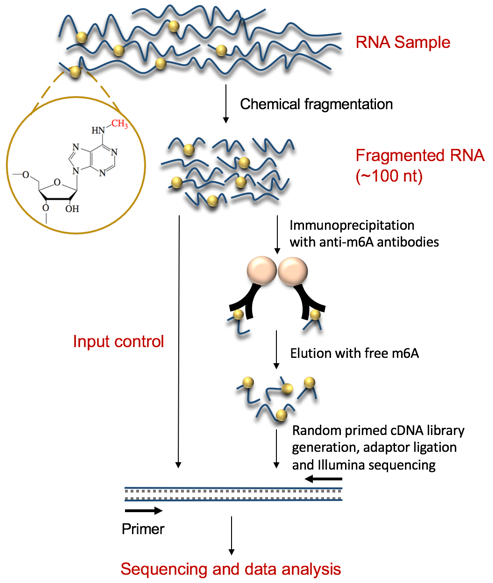
Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing | Nature Protocols
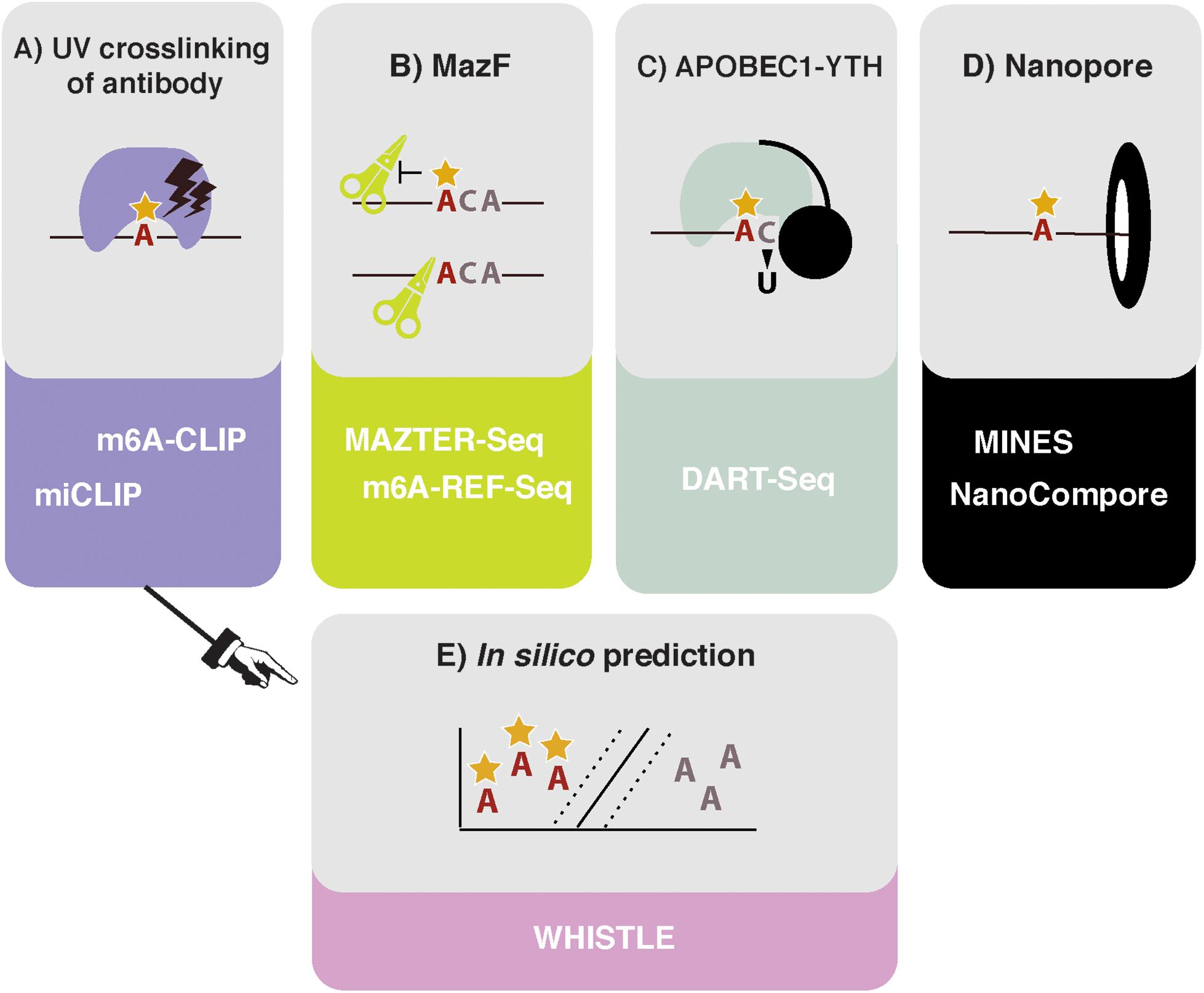
Frontiers | How Do You Identify m6 A Methylation in Transcriptomes at High Resolution? A Comparison of Recent Datasets
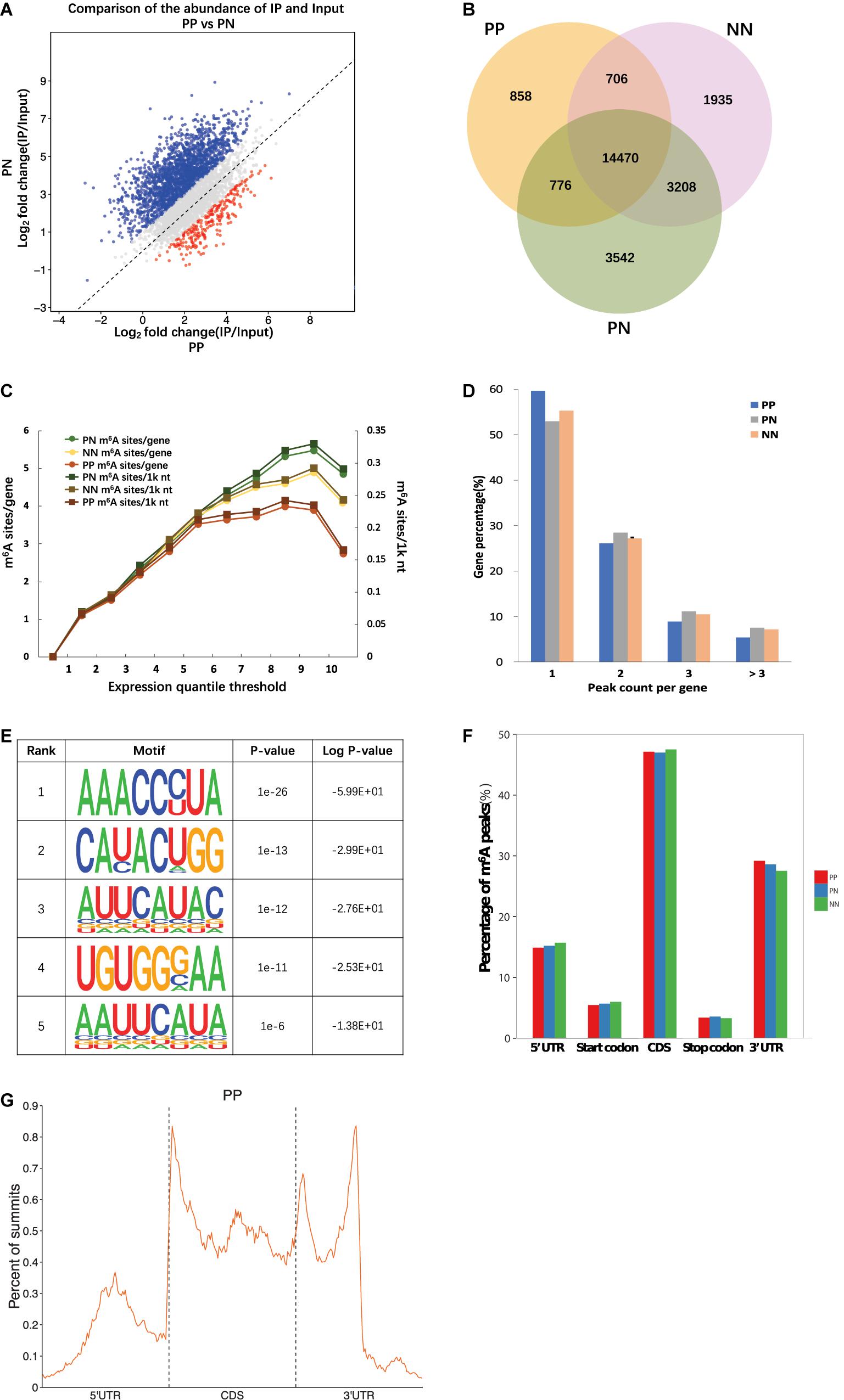
Frontiers | Transcriptome-Wide m6A Methylation in Skin Lesions From Patients With Psoriasis Vulgaris