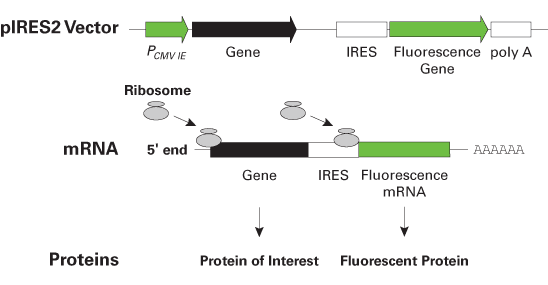
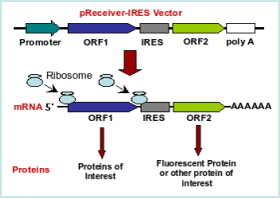
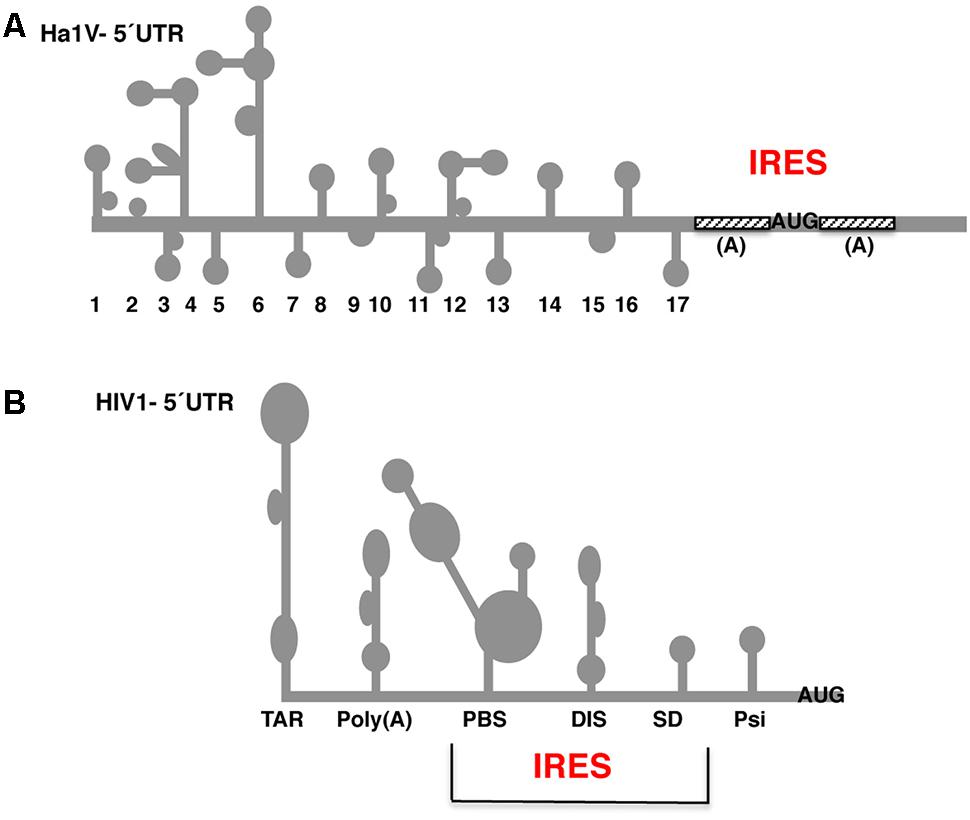
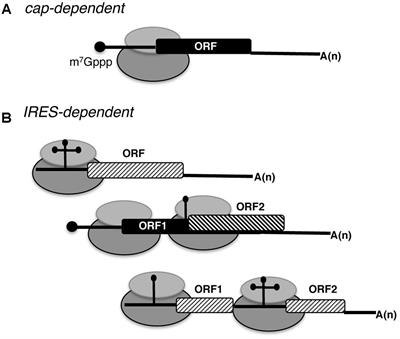
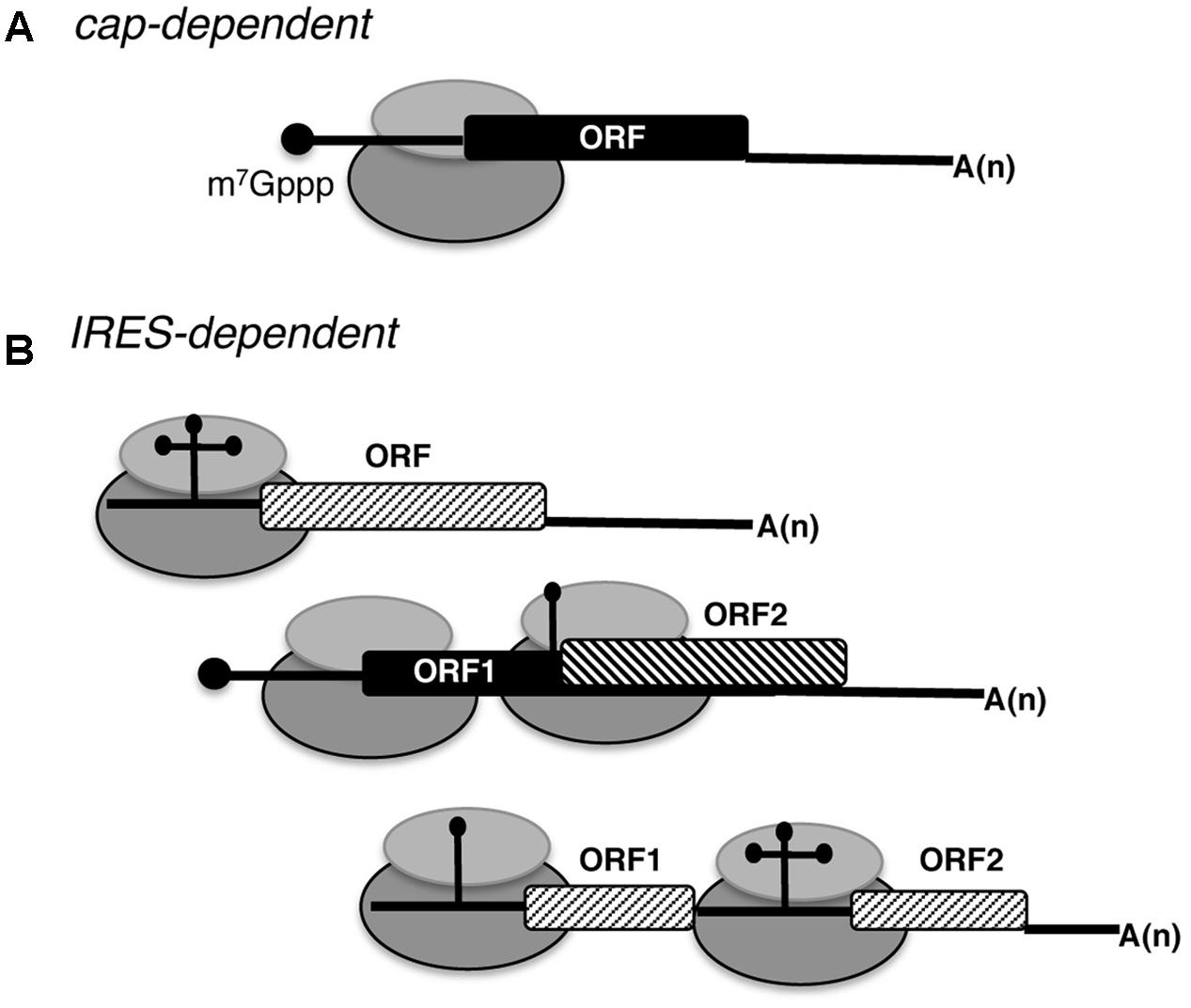
A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity | PNAS

Internal Ribosome Entry Site-mediated Translation of a Mammalian mRNA Is Regulated by Amino Acid Availability* - Journal of Biological Chemistry

Translational Induction of VEGF Internal Ribosome Entry Site Elements During the Early Response to Ischemic Stress | Circulation Research

The IRES element type I. Each of the sequence motifs characteristic for... | Download Scientific Diagram

IRESbase: a Comprehensive Database of Experimentally Validated Internal Ribosome Entry Sites | bioRxiv

Functional characterization of the EMCV IRES in plants - Urwin - 2000 - The Plant Journal - Wiley Online Library
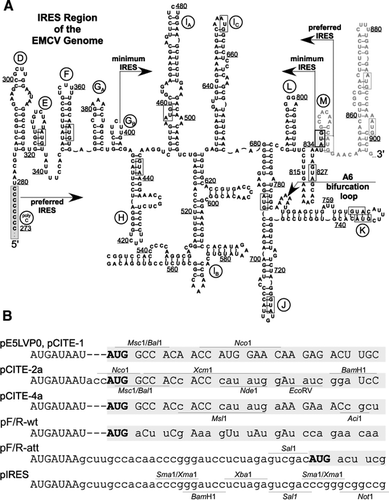
Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location | BioTechniques
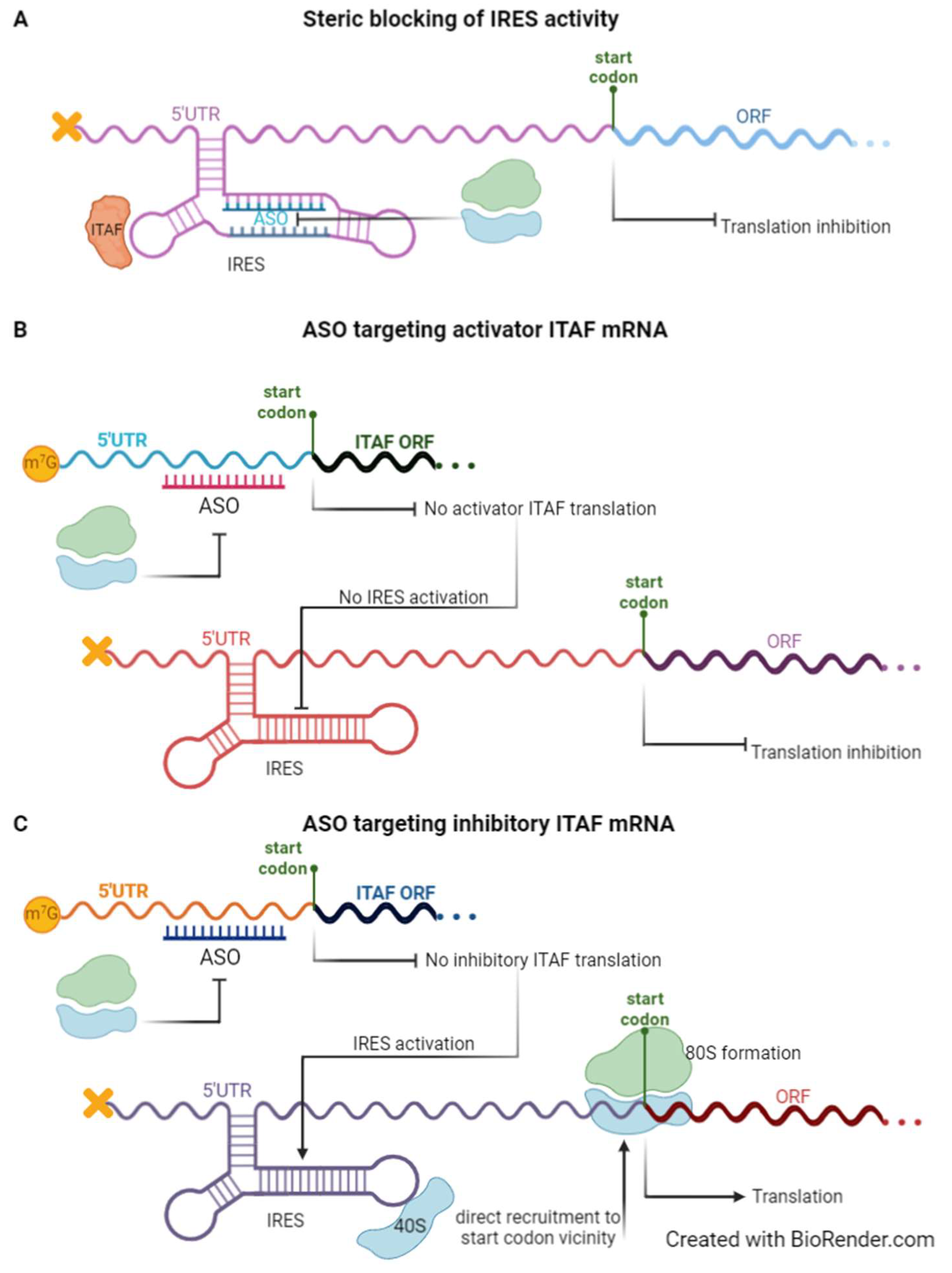
Biomedicines | Free Full-Text | Internal Ribosome Entry Site (IRES)-Mediated Translation and Its Potential for Novel mRNA-Based Therapy Development
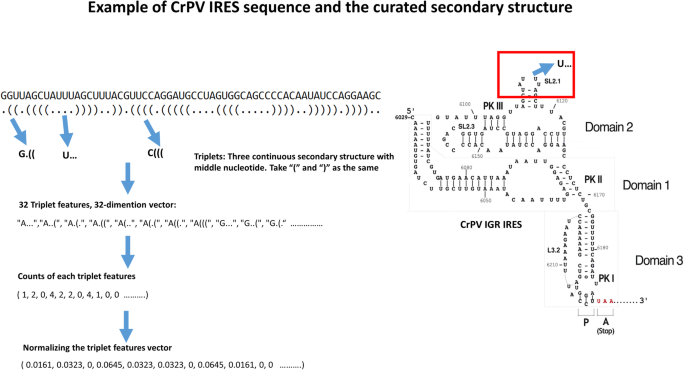
IRESpy: an XGBoost model for prediction of internal ribosome entry sites | BMC Bioinformatics | Full Text
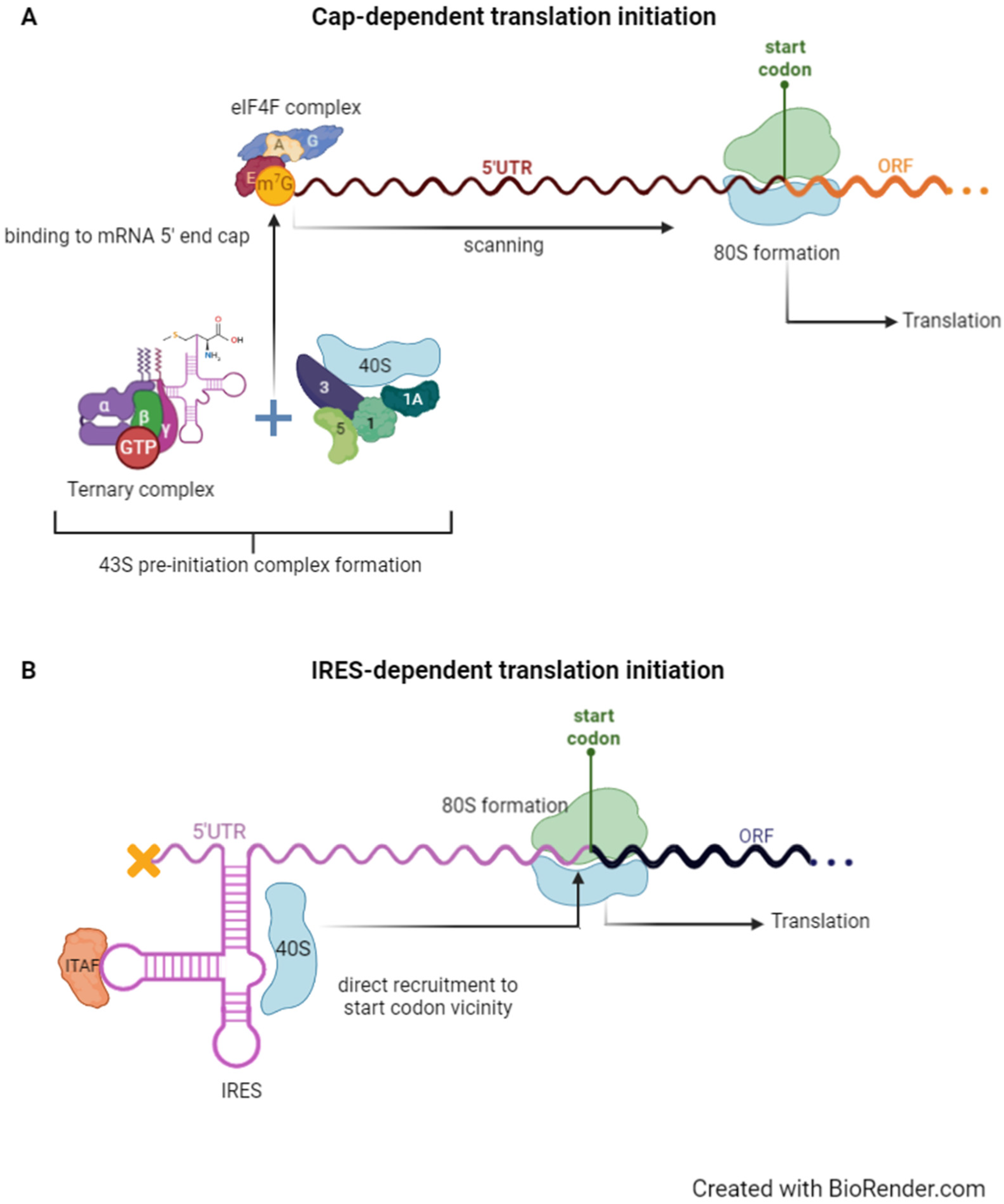
Biomedicines | Free Full-Text | Internal Ribosome Entry Site (IRES)-Mediated Translation and Its Potential for Novel mRNA-Based Therapy Development