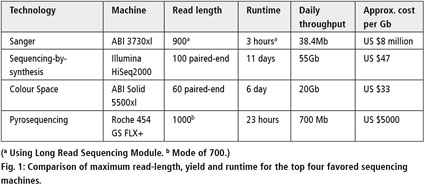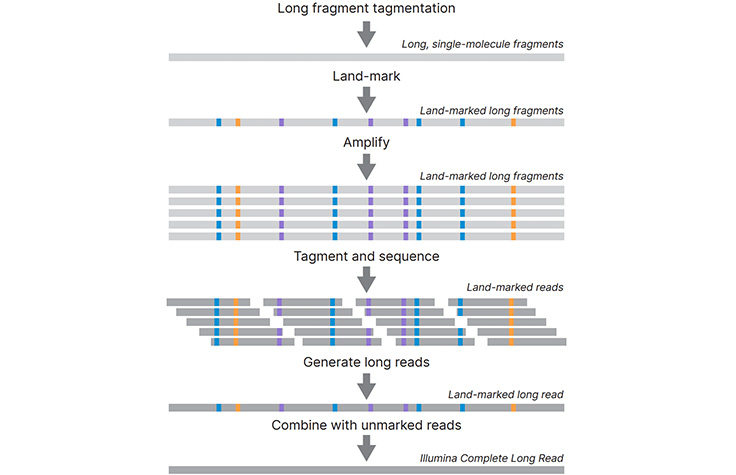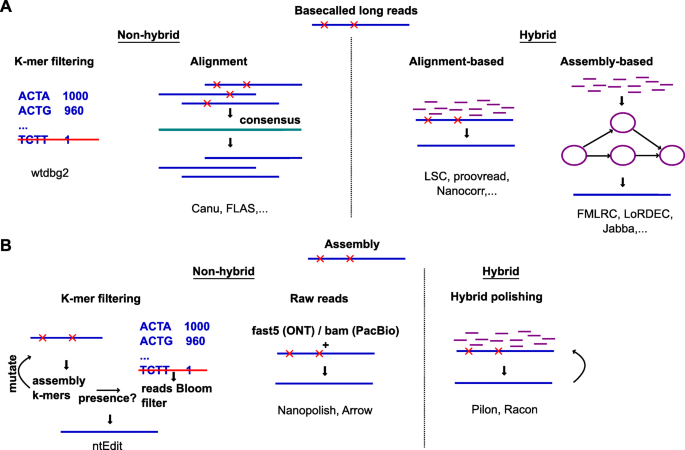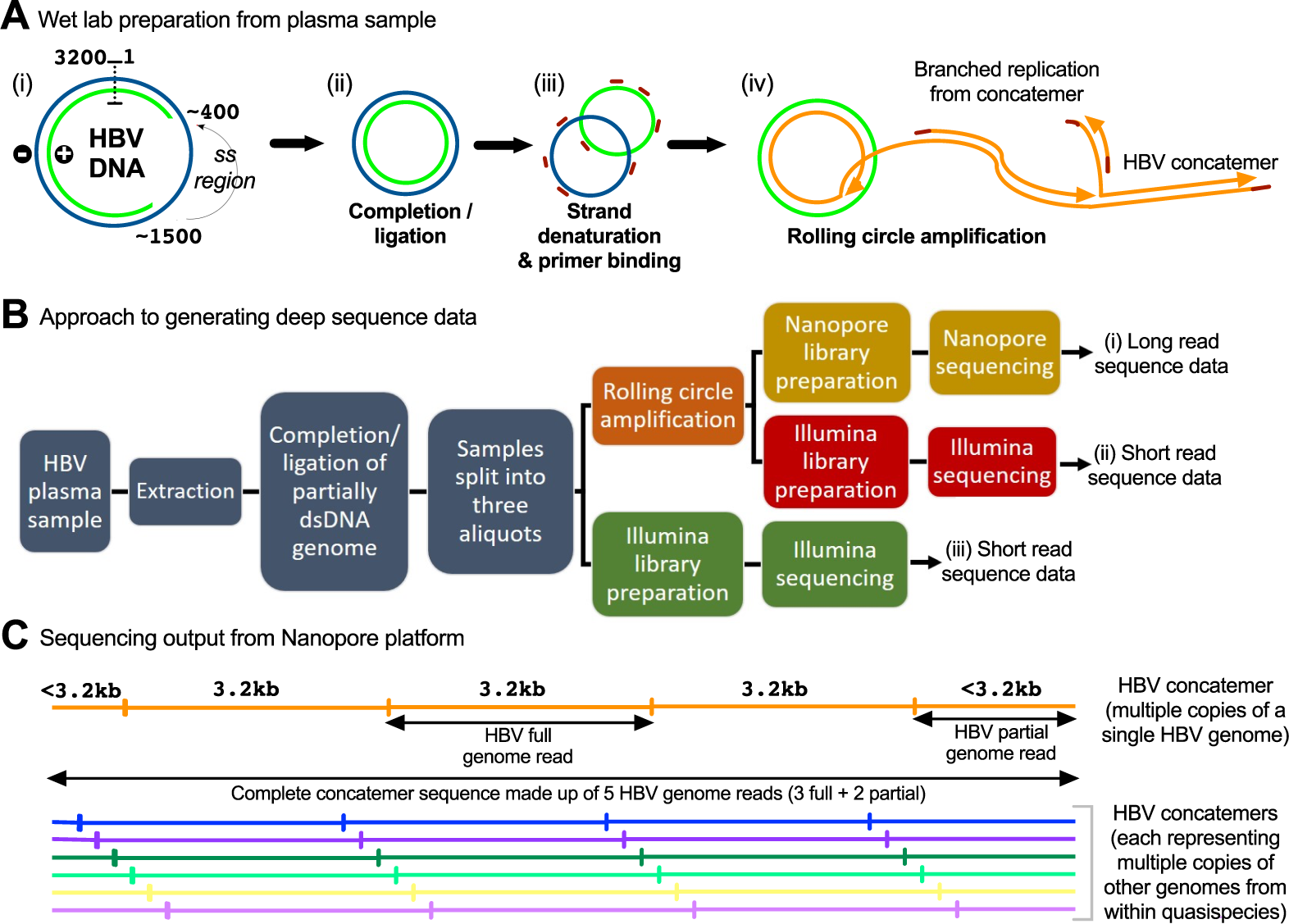
Illumina and Nanopore methods for whole genome sequencing of hepatitis B virus (HBV) | Scientific Reports

The distribution of read lengths in 454, Illumina and PacBio budgerigar... | Download Scientific Diagram

Histogram of read variants' lengths in Illumina sequencing data. The... | Download Scientific Diagram