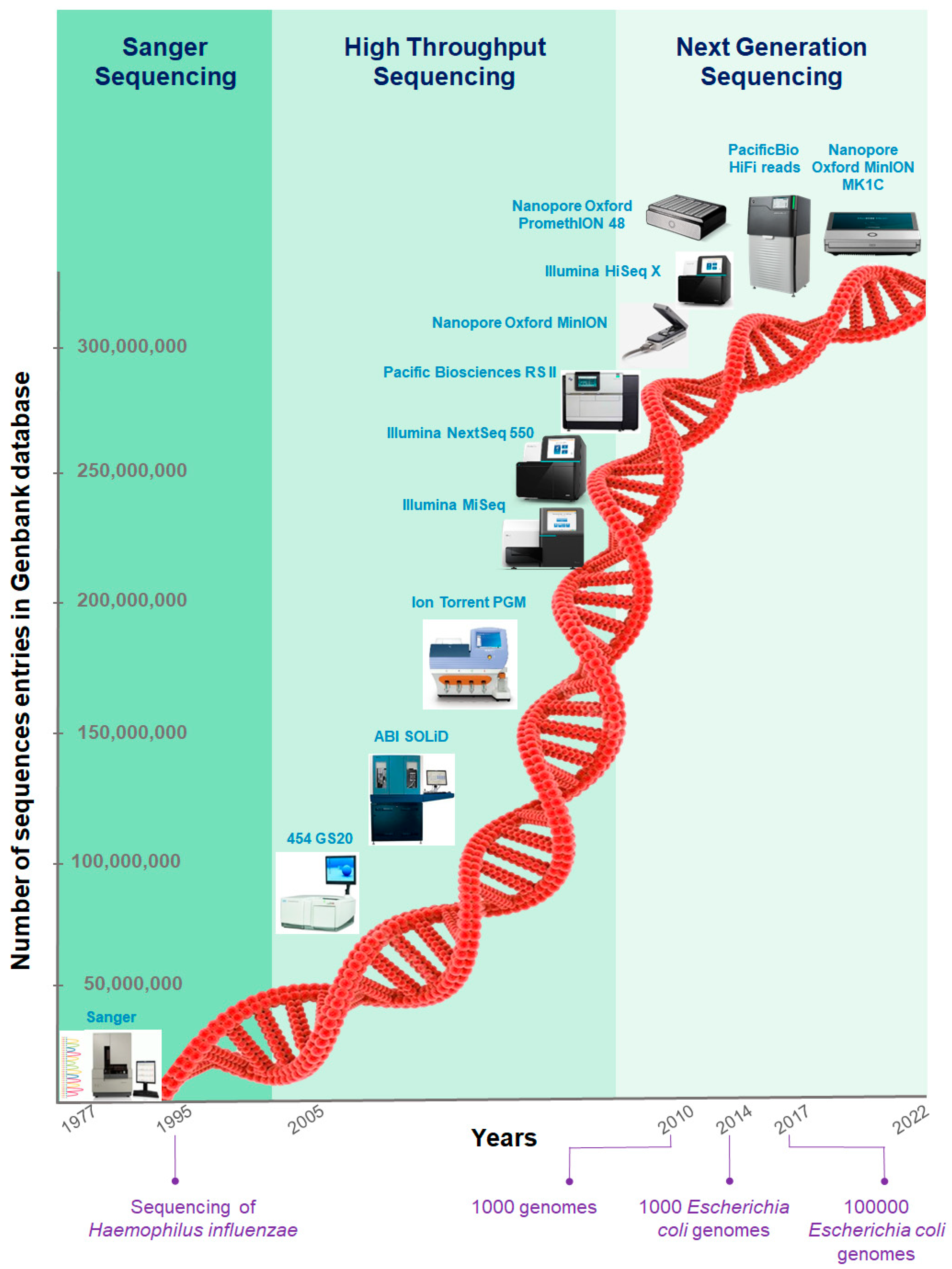
IJMS | Free Full-Text | Application and Challenge of 3rd Generation Sequencing for Clinical Bacterial Studies

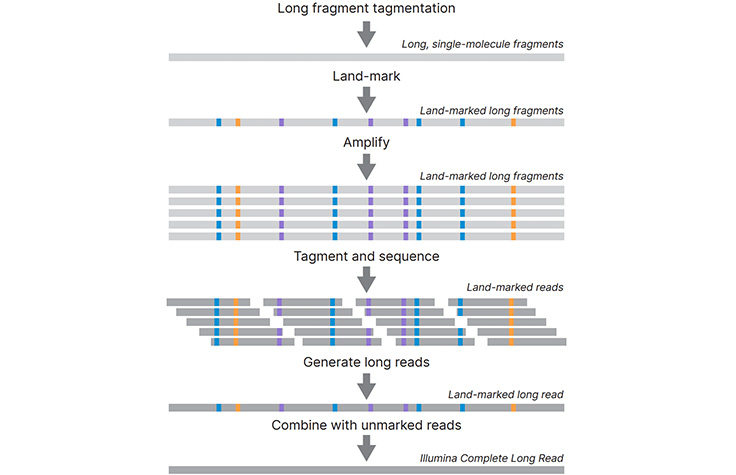
Illumina HiSeq v4 Sequencing Services Yielding 1Tb Data / Week Now Available on Genohub | Genohub Blog
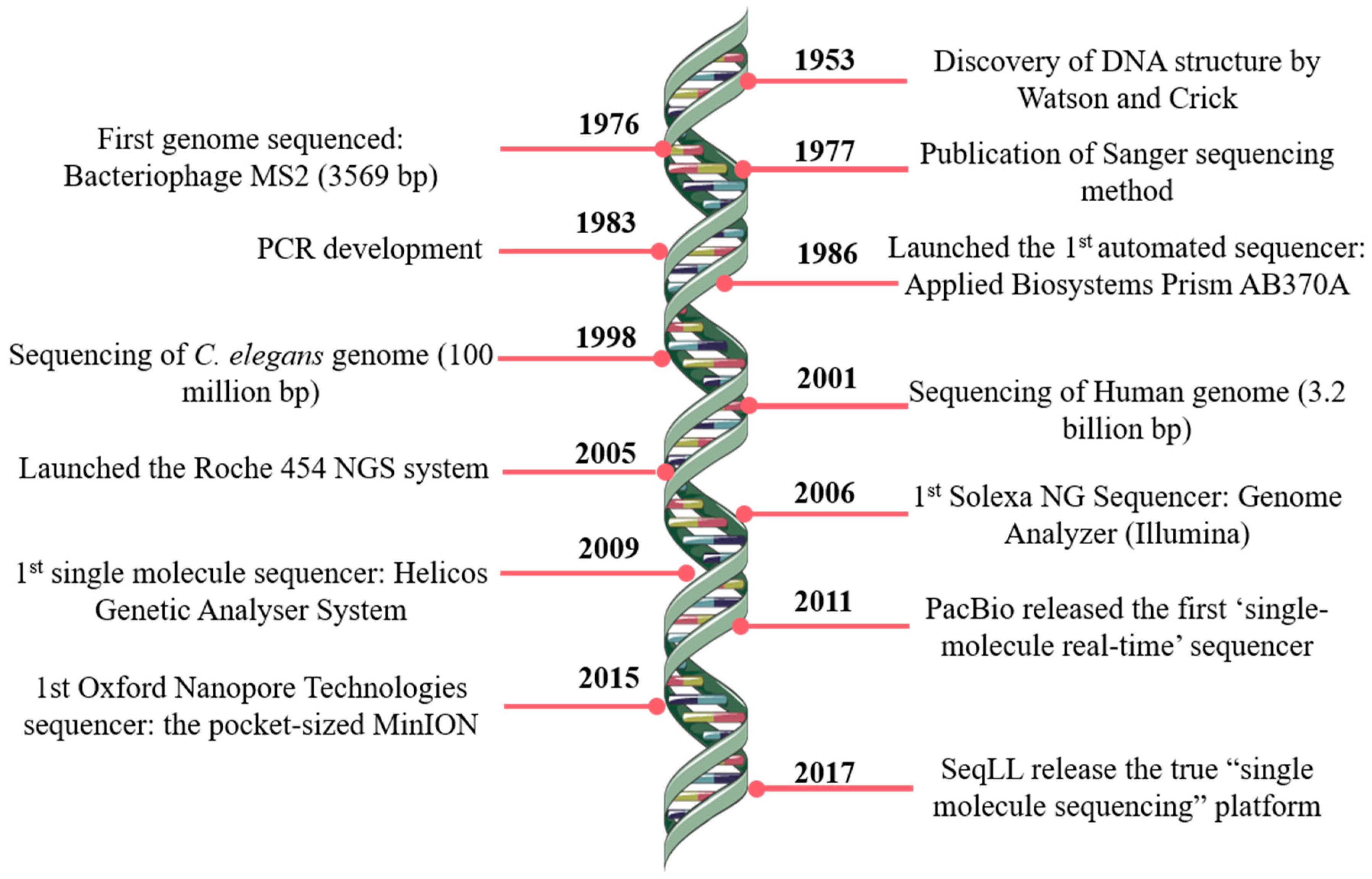
JCM | Free Full-Text | Bioinformatics and Computational Tools for Next-Generation Sequencing Analysis in Clinical Genetics
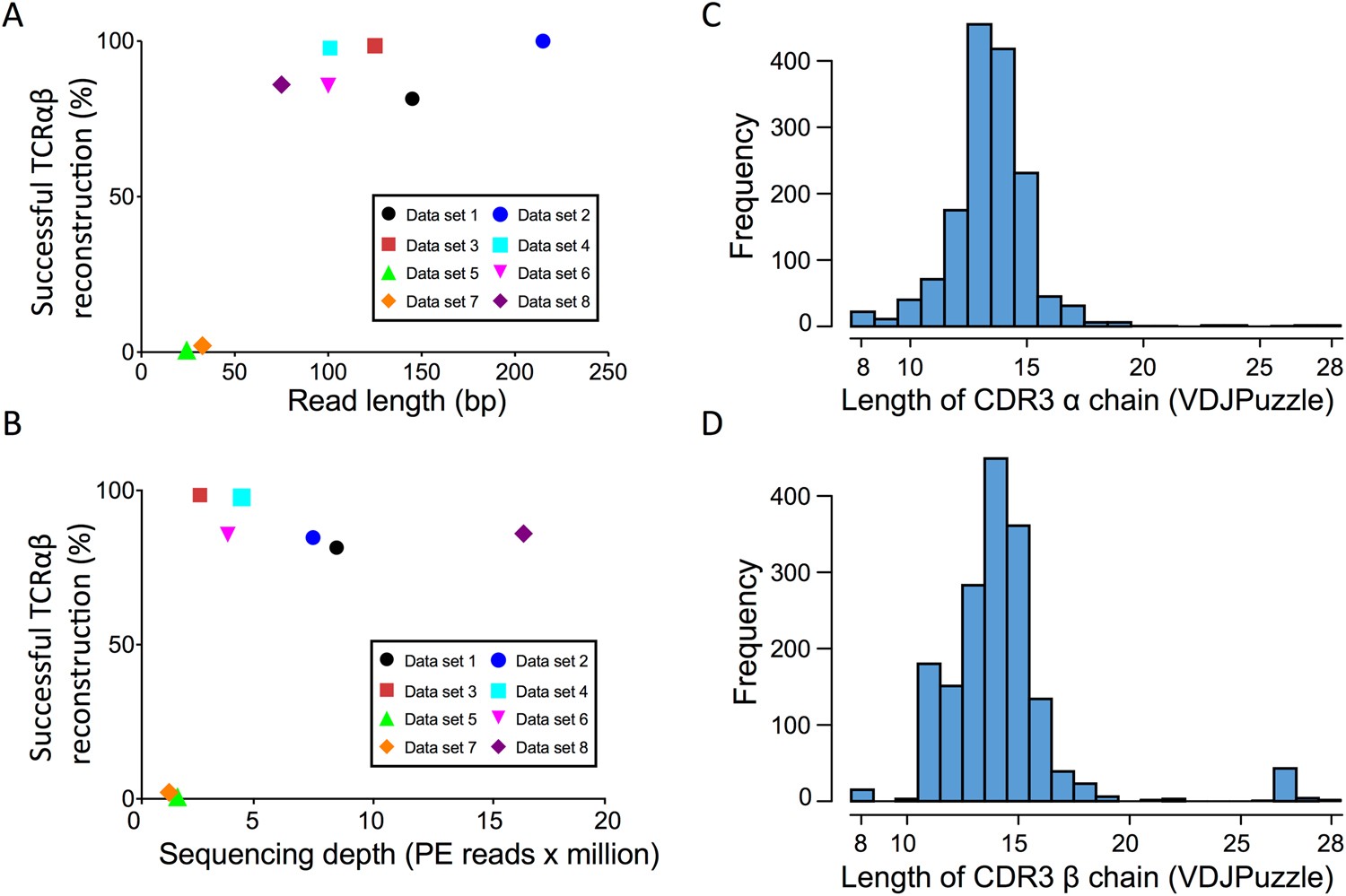
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

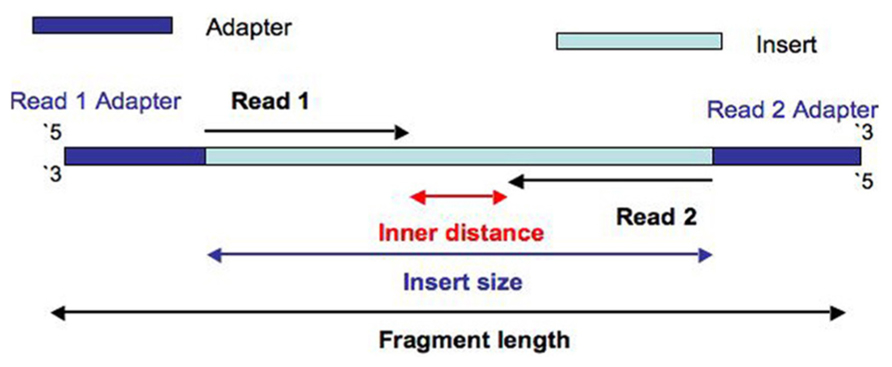
Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge
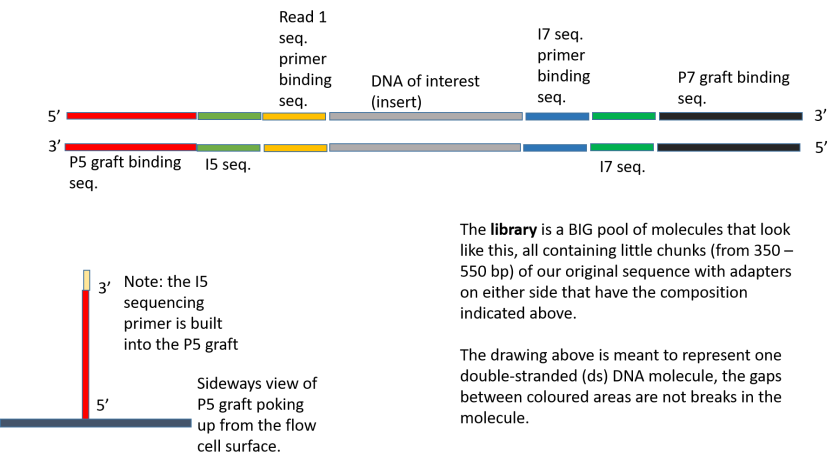
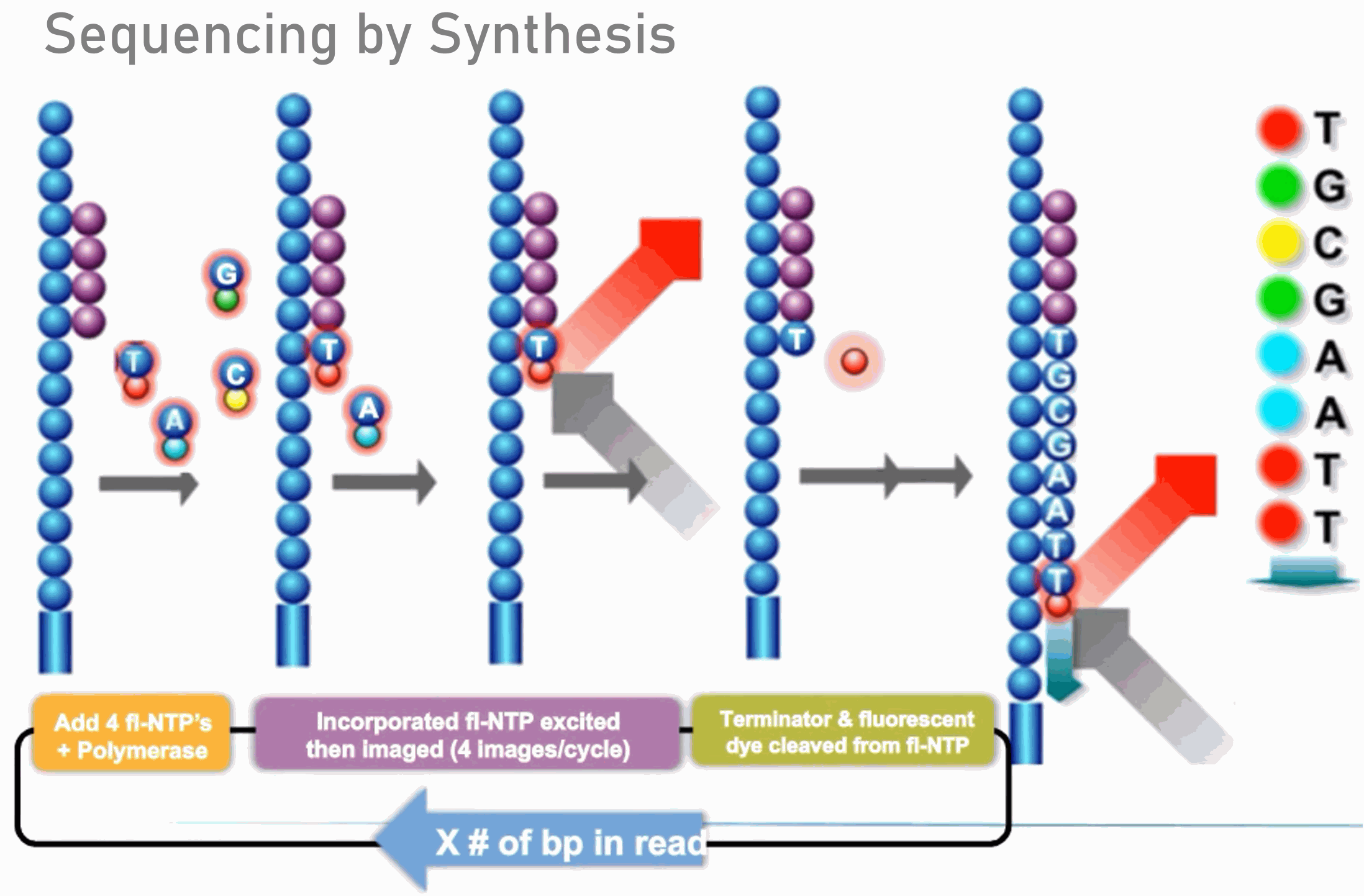
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics
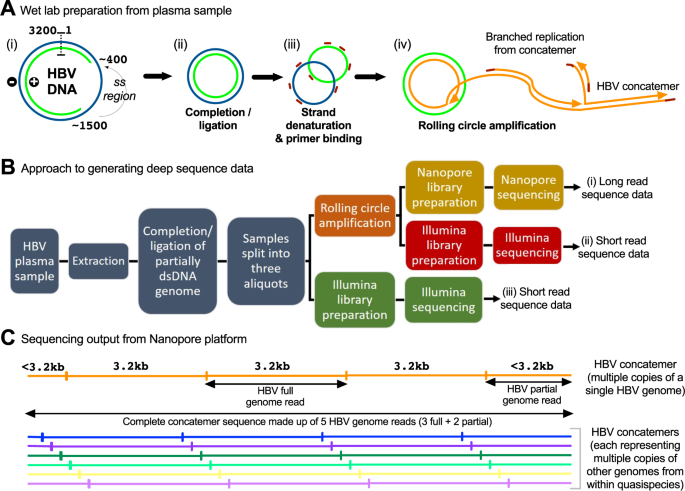
Illumina and Nanopore methods for whole genome sequencing of hepatitis B virus (HBV) | Scientific Reports
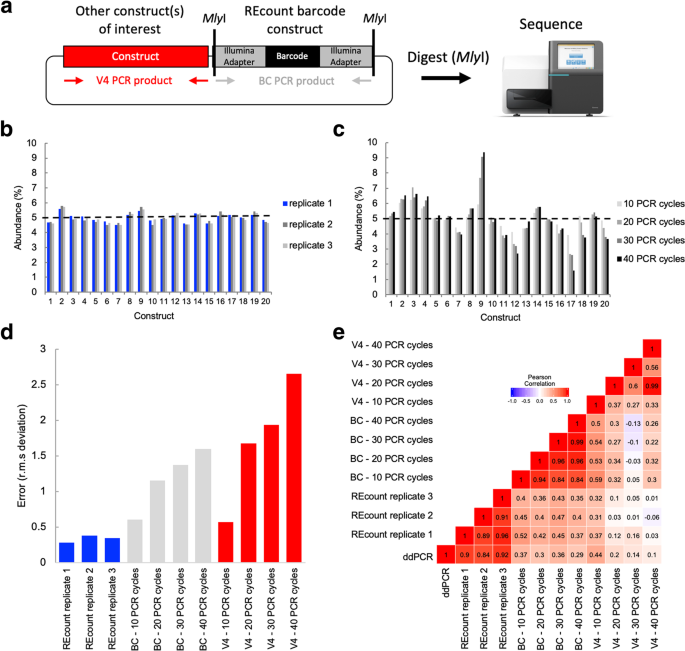
Measuring sequencer size bias using REcount: a novel method for highly accurate Illumina sequencing-based quantification | Genome Biology | Full Text