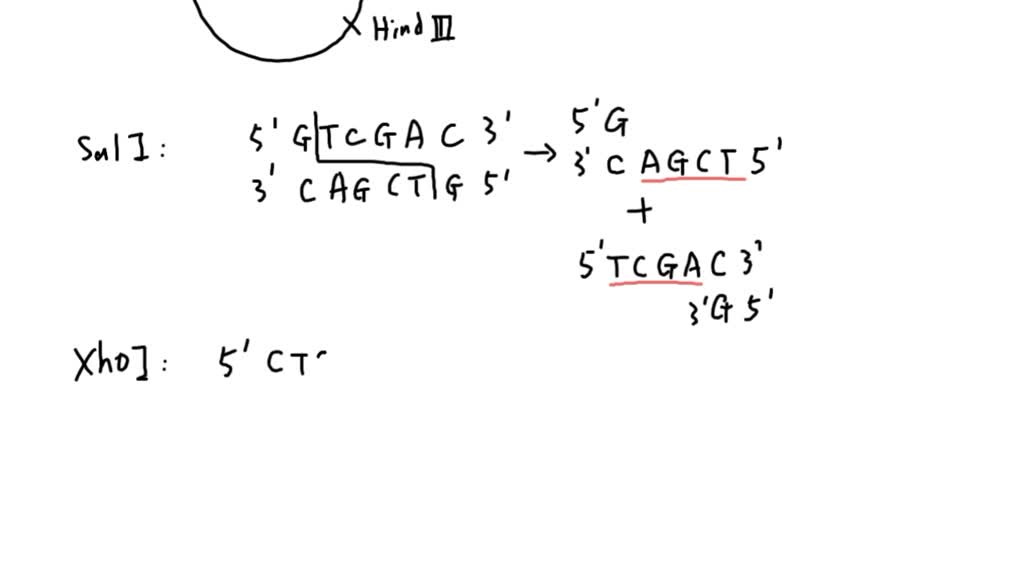
SOLVED: 2. The figure ' below shows plasmid with the sites of cleavage for restriction enzymes marked. The enzymes are Sall, Xhol, EcoRV, and HindIII Their recognition sequences and site of cleavage
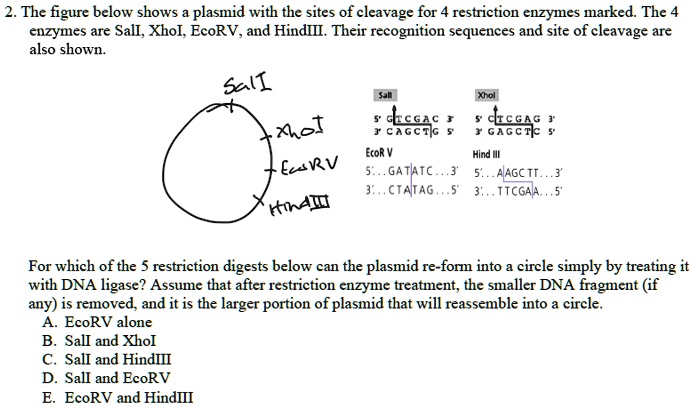
SOLVED: 2. The figure ' below shows plasmid with the sites of cleavage for restriction enzymes marked. The enzymes are Sall, Xhol, EcoRV, and HindIII Their recognition sequences and site of cleavage
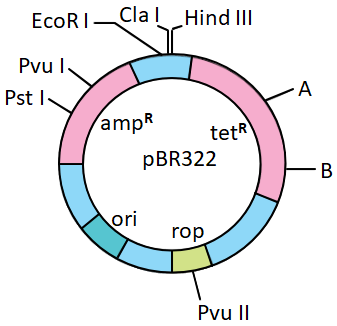
A and B in the pBR 322, shown in the diagram given below, respectively represent recognition sequences of: 1. BamH I and Sma I 2. Hind II and Sma I 3. BamH

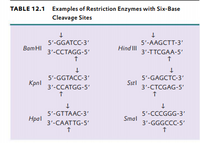
The restriction enzyme responsible for the cleavage of following sequence is 5' - G - T - C - G - A - C - 3'3' - C - A - G - C - T - G - 5'

Restriction Enzymes/endonucleases Restriction enzymes are proteins that scan the DNA for specific sequences, usually palindromic sequences of 4 to 8 nucleotides, - ppt download

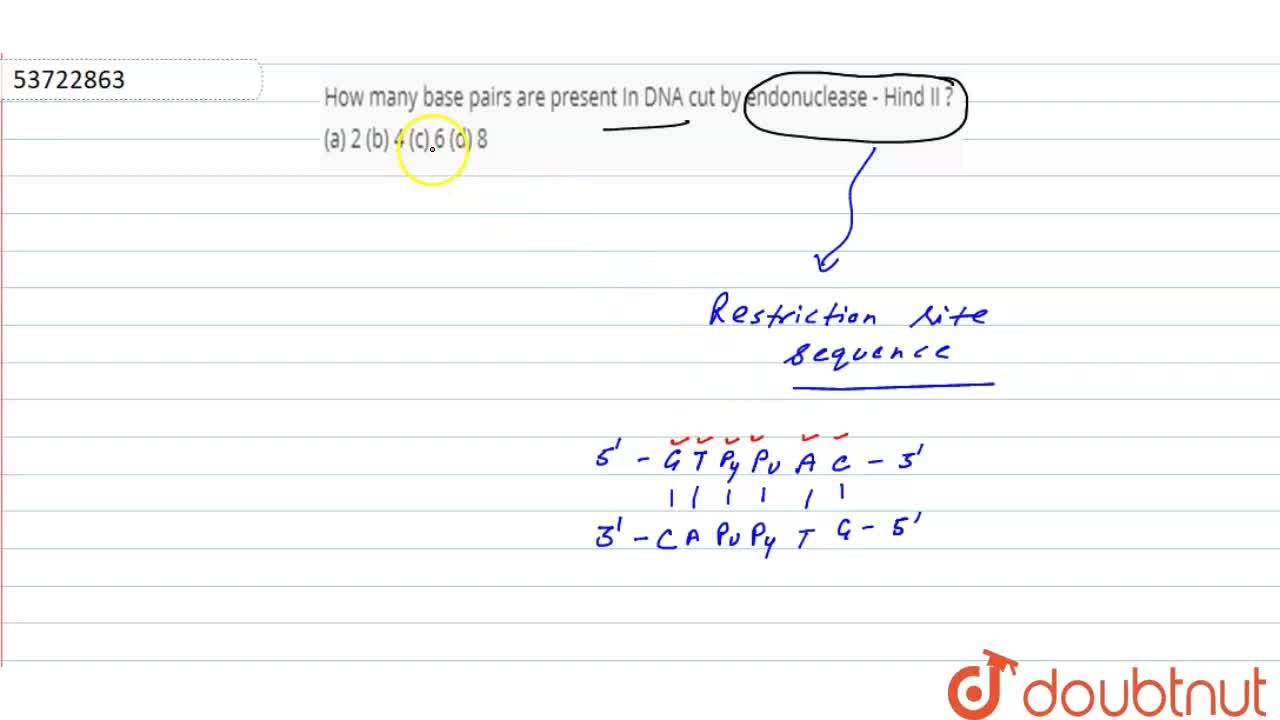
Assertion : The first restriction endonuclease was Hine IIReason :Hind II always cut DNA molecul... - YouTube

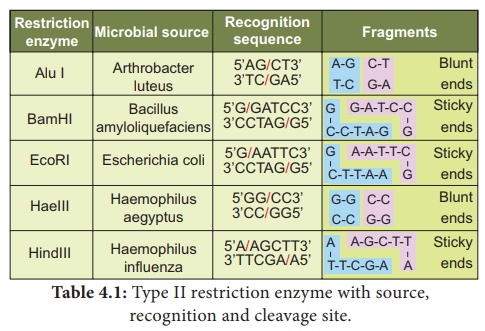
Recognition sequences Restriction enzyme EcoRI cuts the DNA into fragments. Sticky end. - ppt download

Restriction endonuclease - Hind II always cuts DNA molecules at a particular point by recognizin... - YouTube
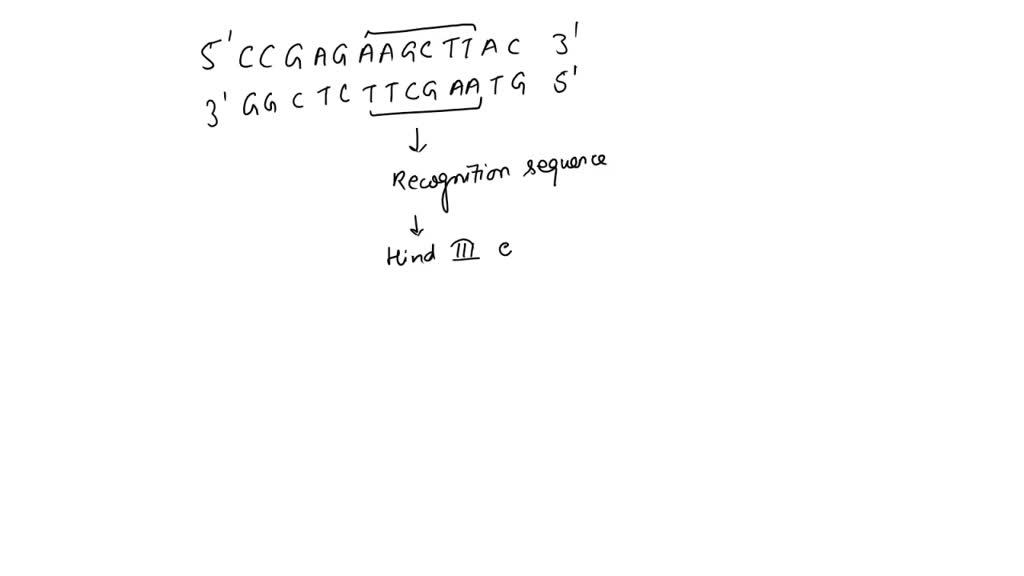
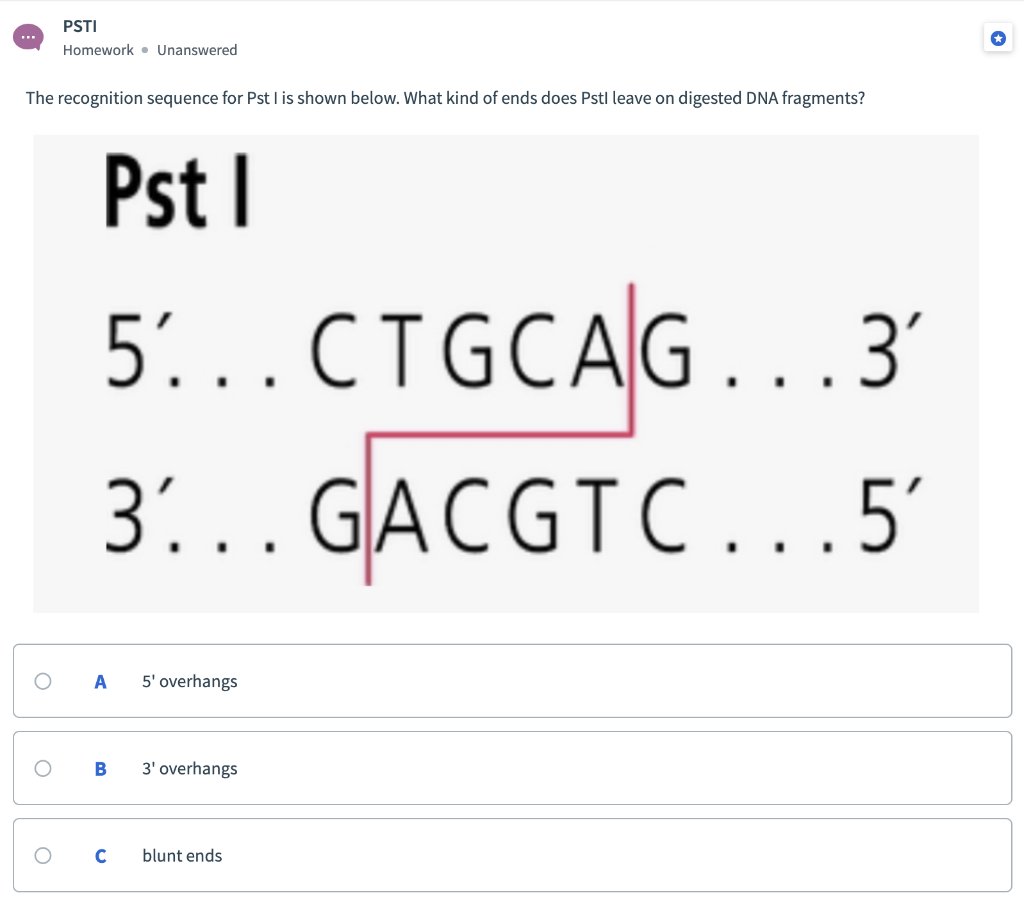
SOLVED: The DNA sequence (5 '-CCGAGAGATCTAC-3') contains a six-base sequence that is a recognition and cutting site for a restriction enzyme. ( 4 points) (a) What is this sequence? (2 points) (b)

It was found that Hind 2 always cuts DNA molecules at a particular point by recognising a specific sequence - Brainly.in