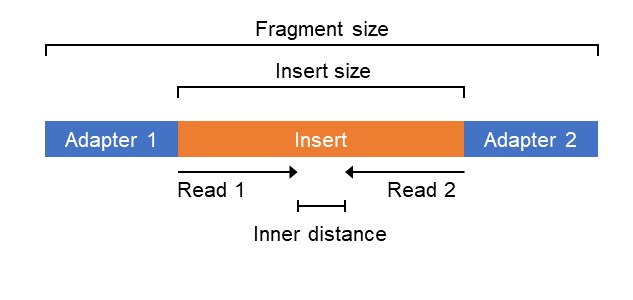
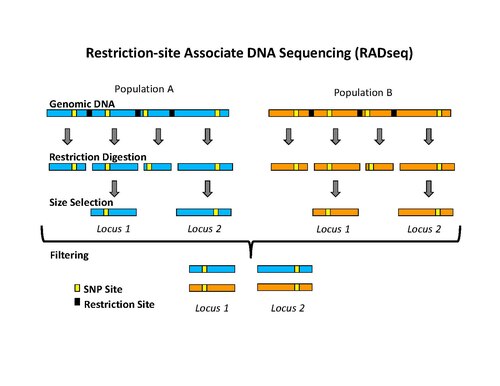
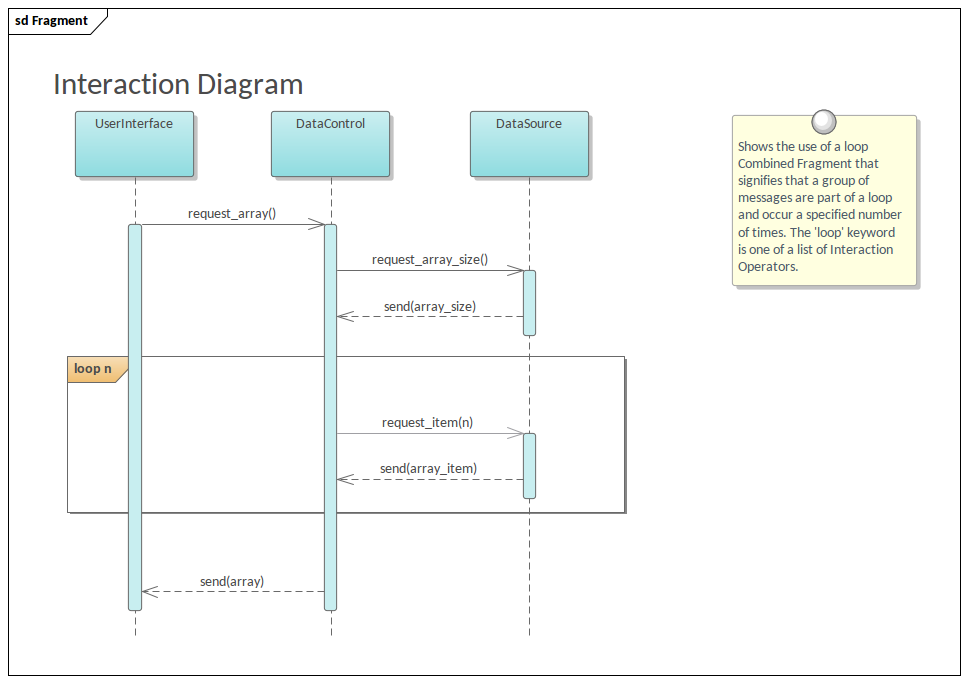
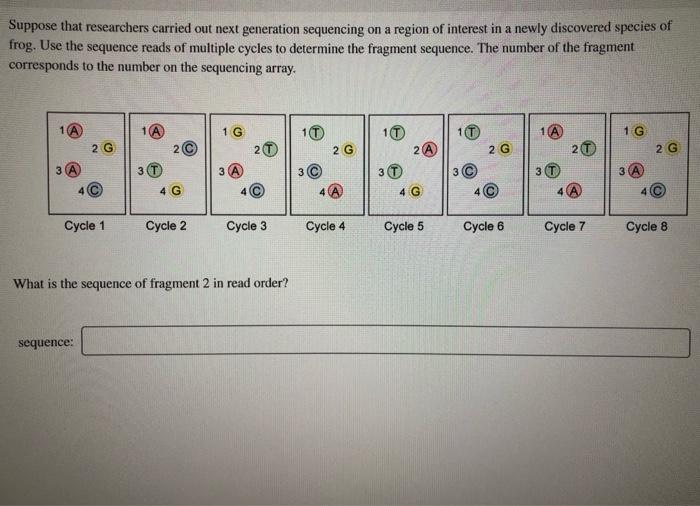
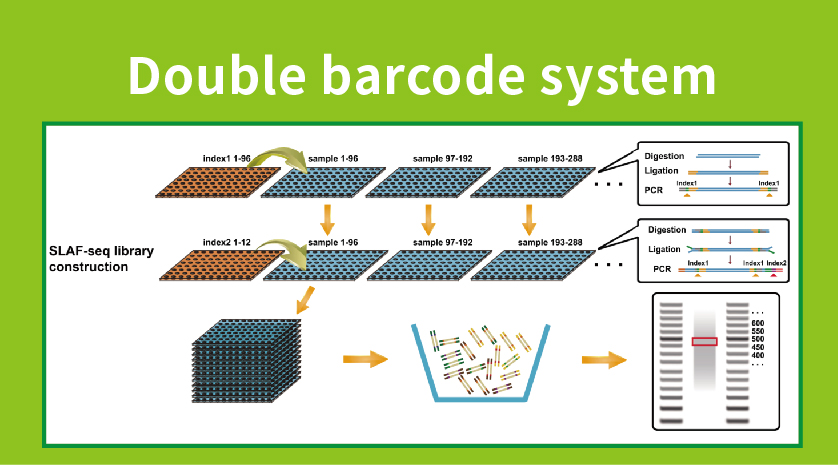
PacBio Sequencing adapters enable the circularization of DNA fragments.... | Download Scientific Diagram

A) Process of SMRT sequencing. A DNA fragment is bluntly ligated to... | Download Scientific Diagram

Next-generation sequencing of Okazaki fragments extracted from Saccharomyces cerevisiae - ScienceDirect

Modeling of RNA-seq fragment sequence bias reduces systematic errors in transcript abundance estimation | Nature Biotechnology

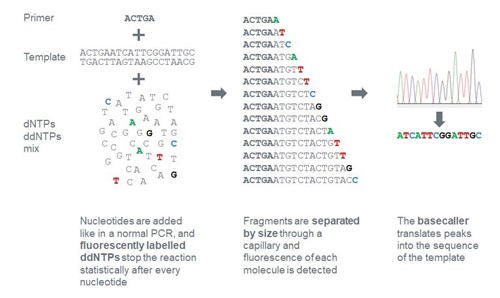
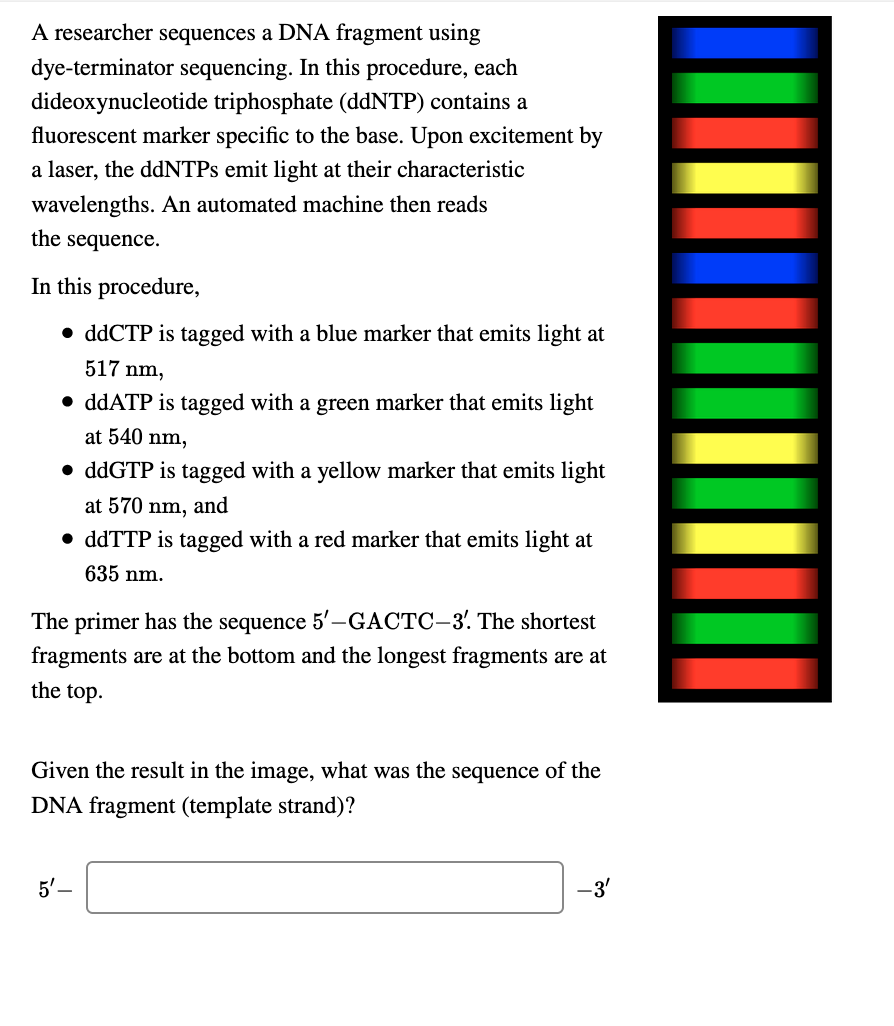
Two common forms of DNA sequencing are shown. The process for one DNA... | Download Scientific Diagram