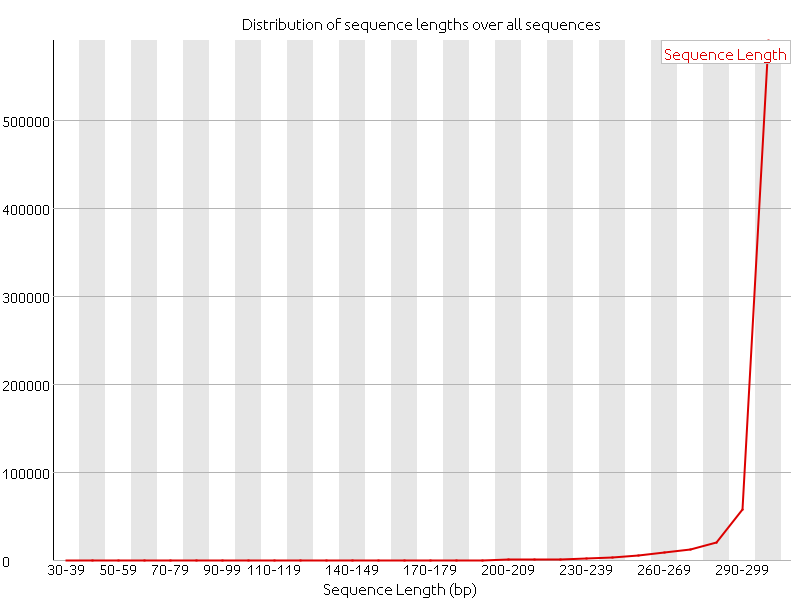
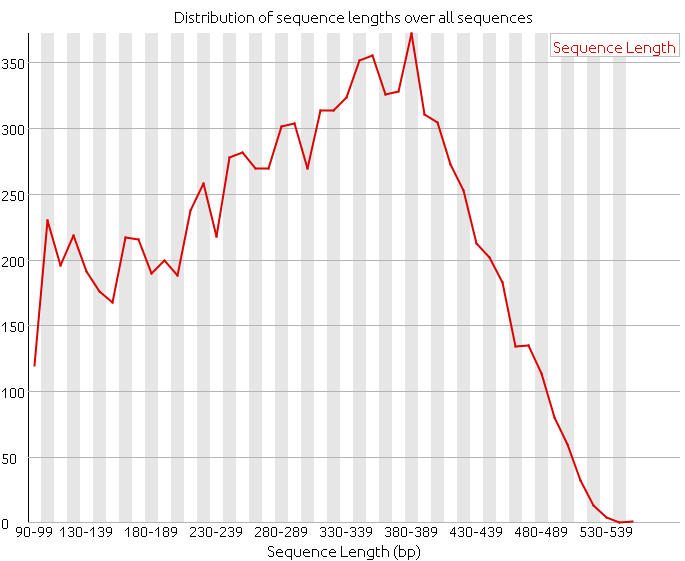
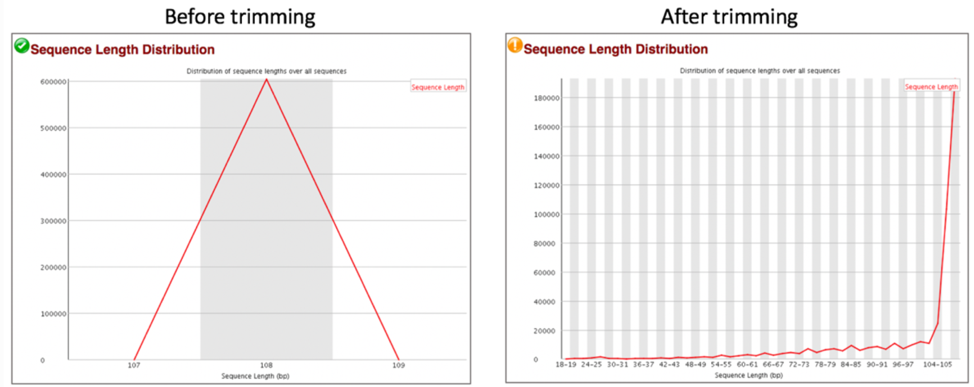
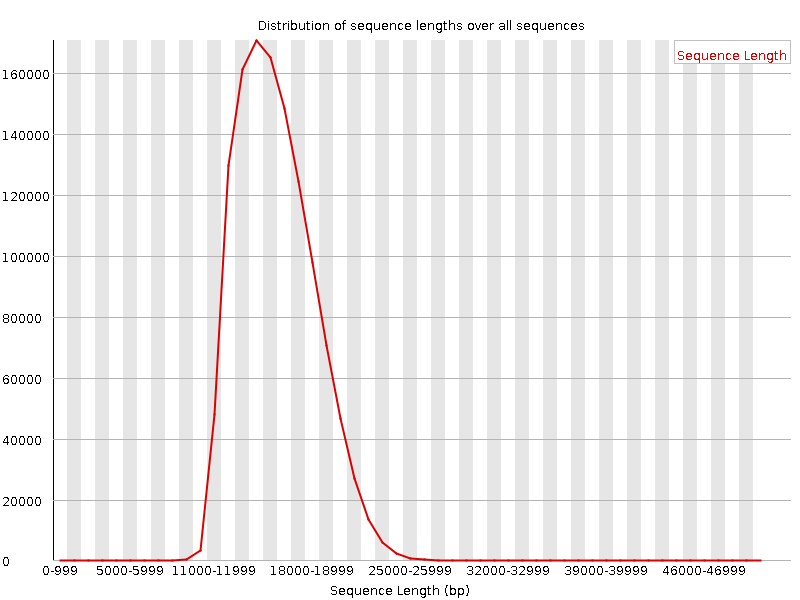
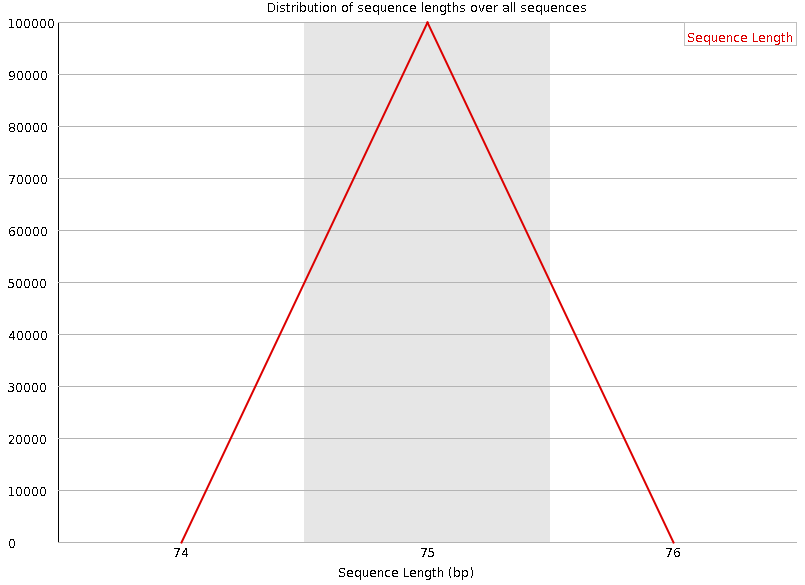
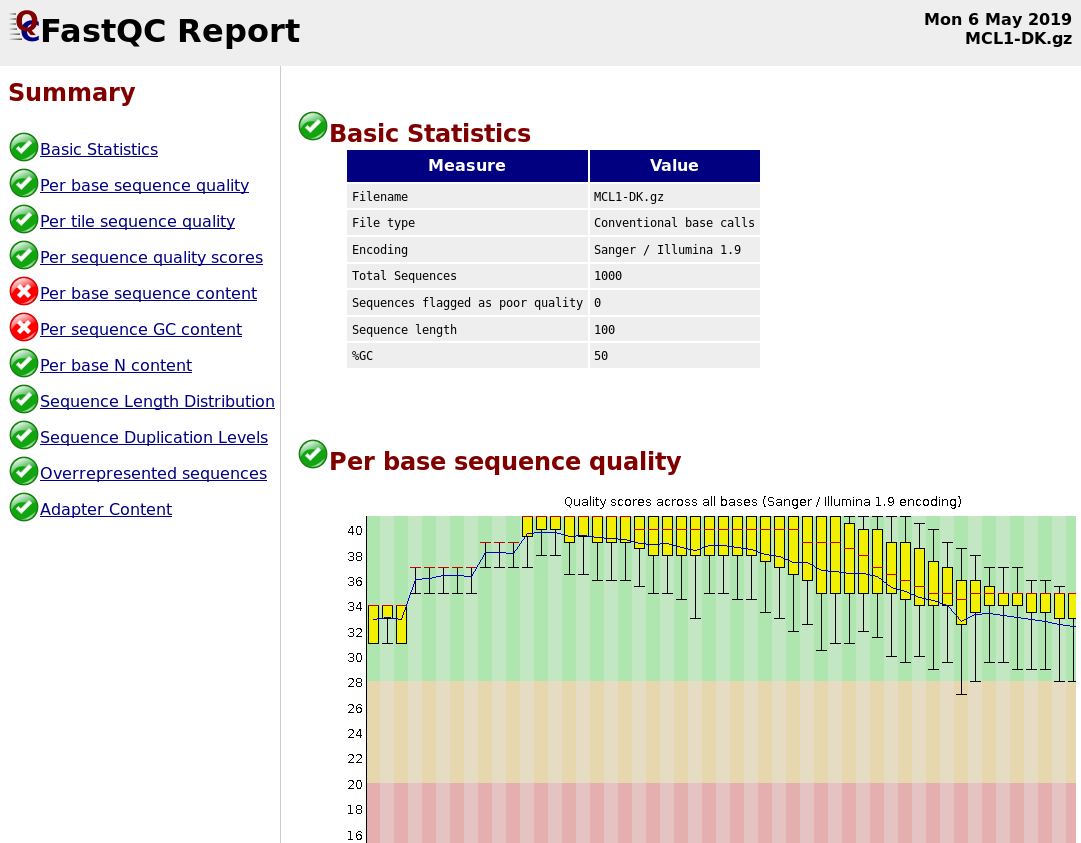
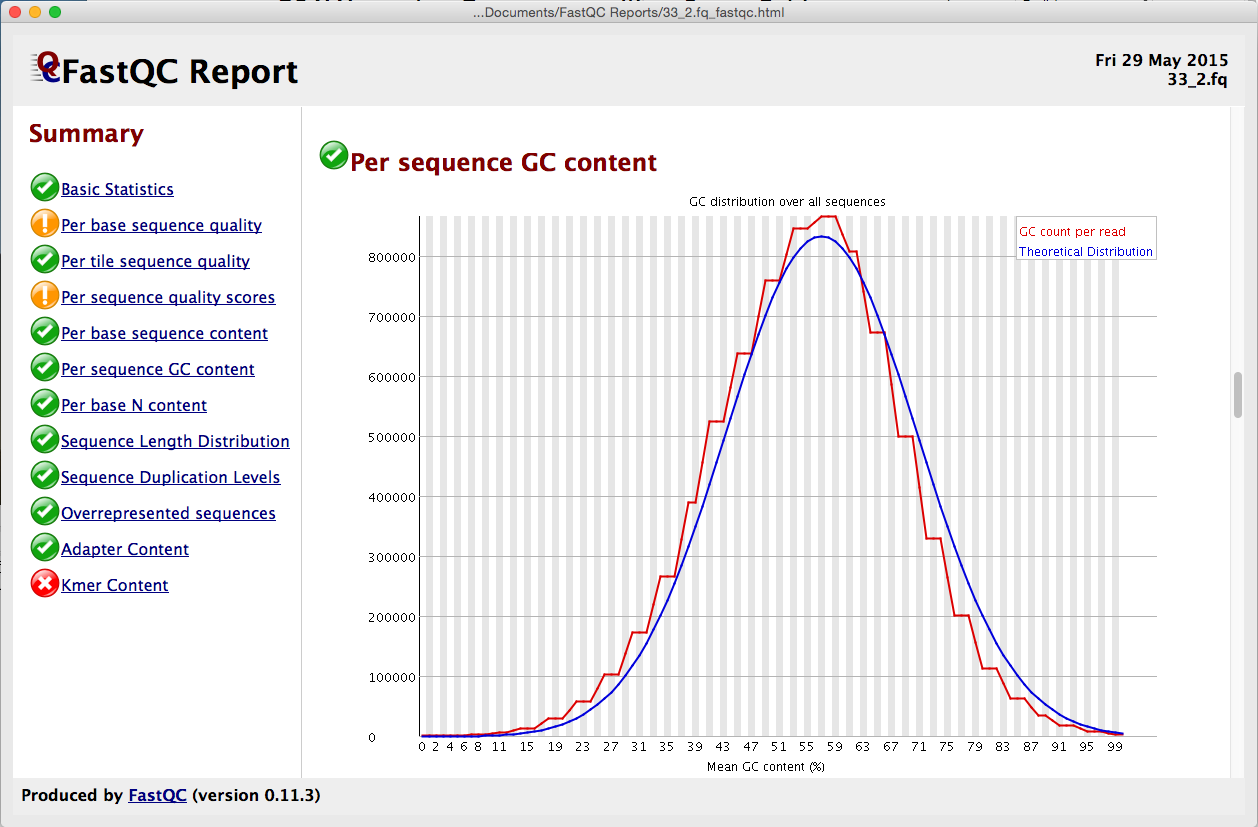
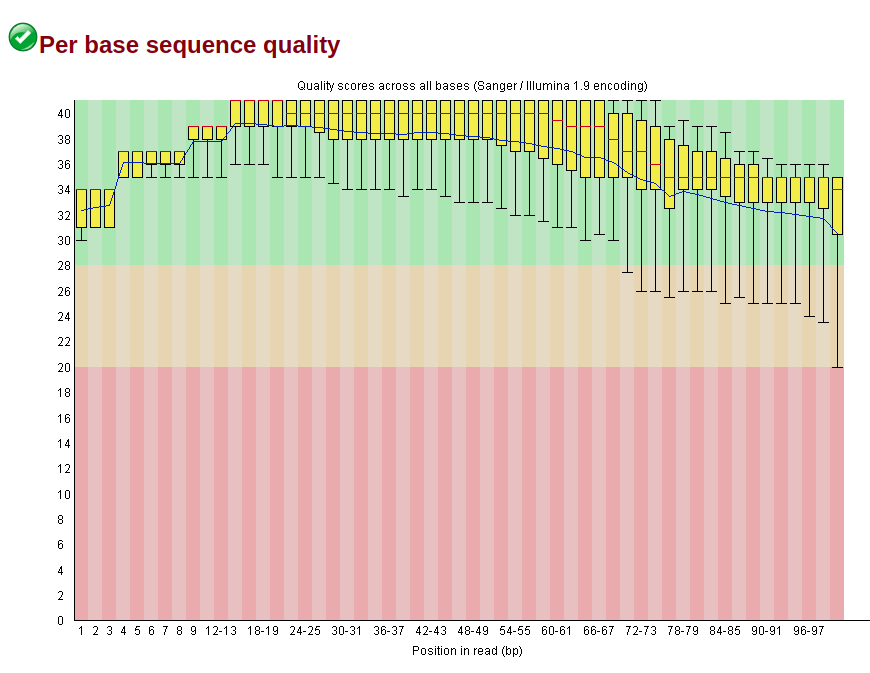
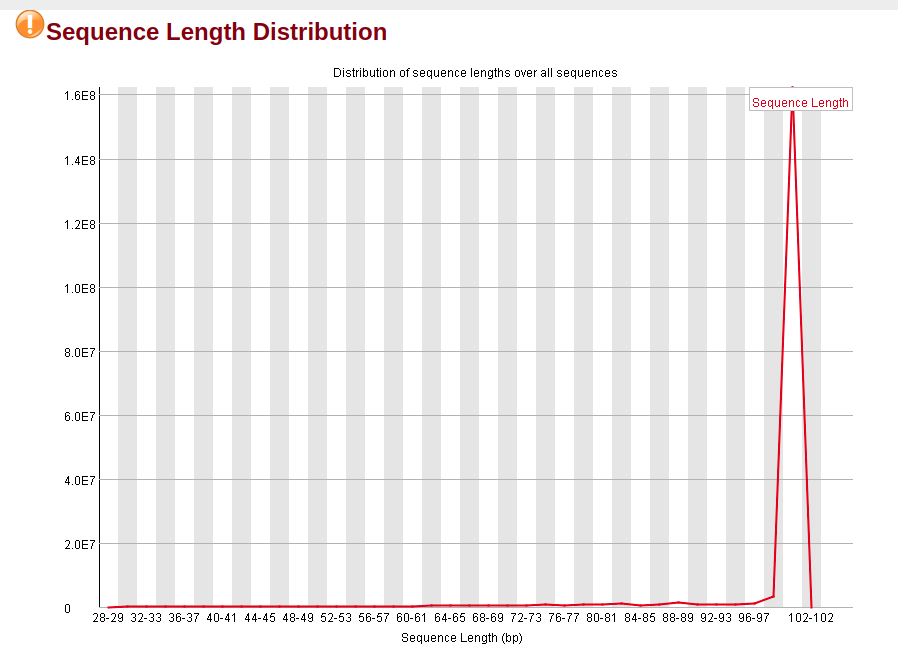
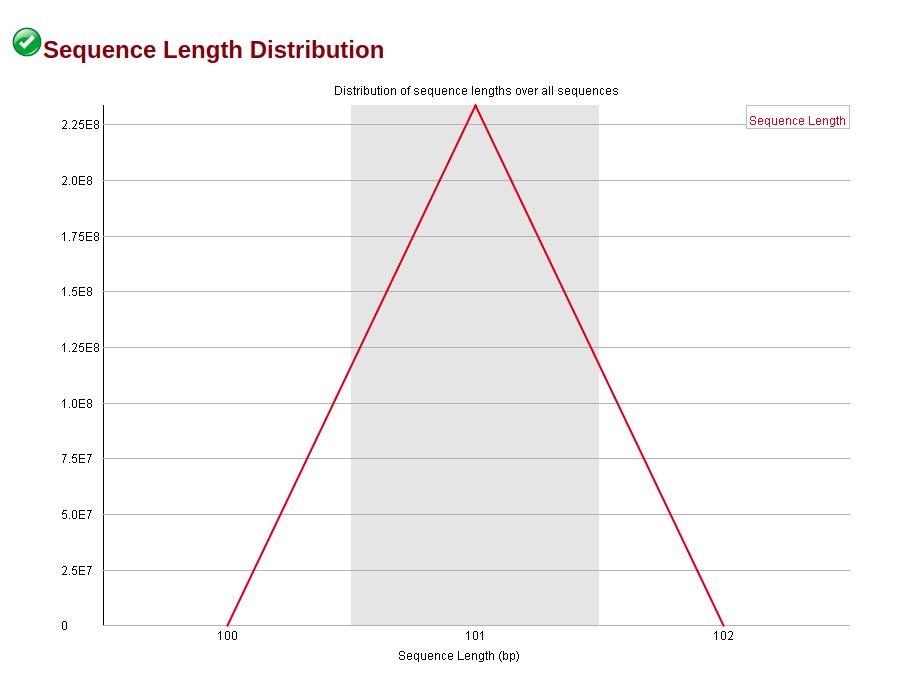
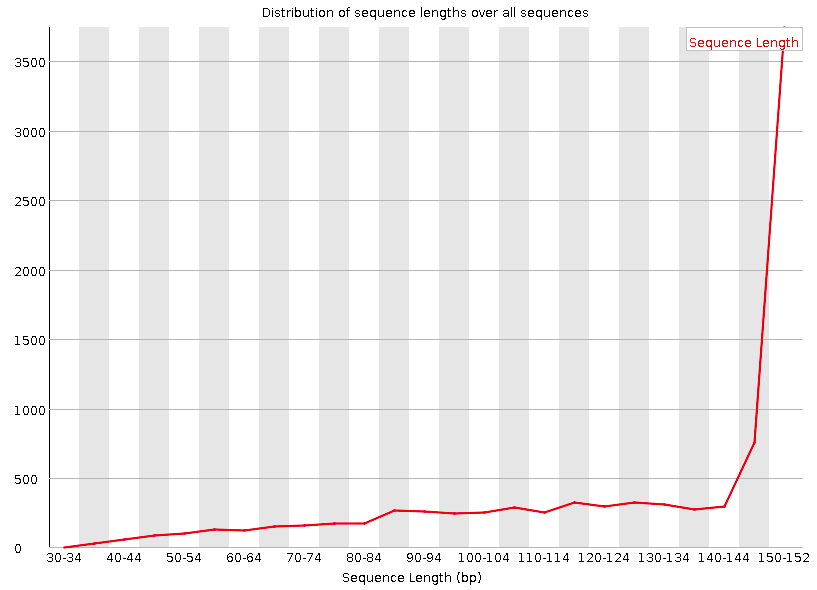
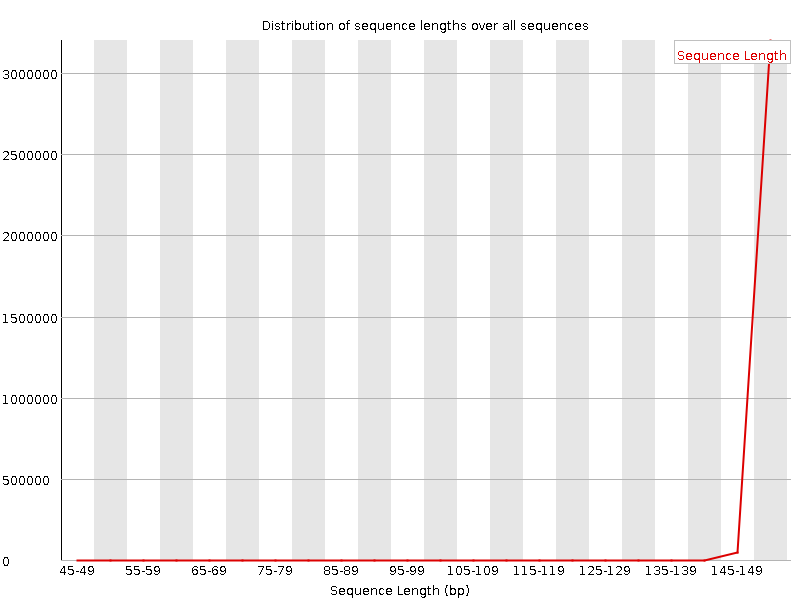
Distribution of Sequence Length After Adapter Removal Using FastQC. The... | Download Scientific Diagram

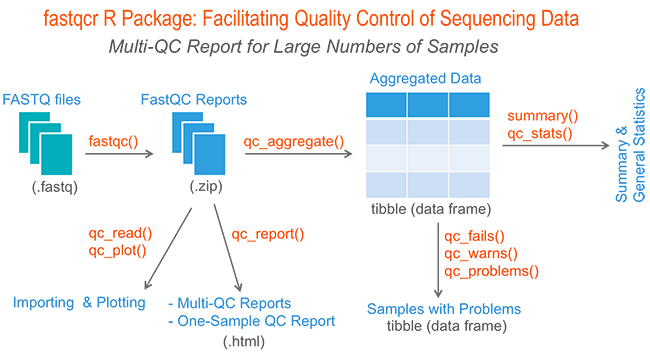
An open-sourced bioinformatic pipeline for the processing of Next-Generation Sequencing derived nucleotide reads: Identification and authentication of ancient metagenomic DNA | bioRxiv