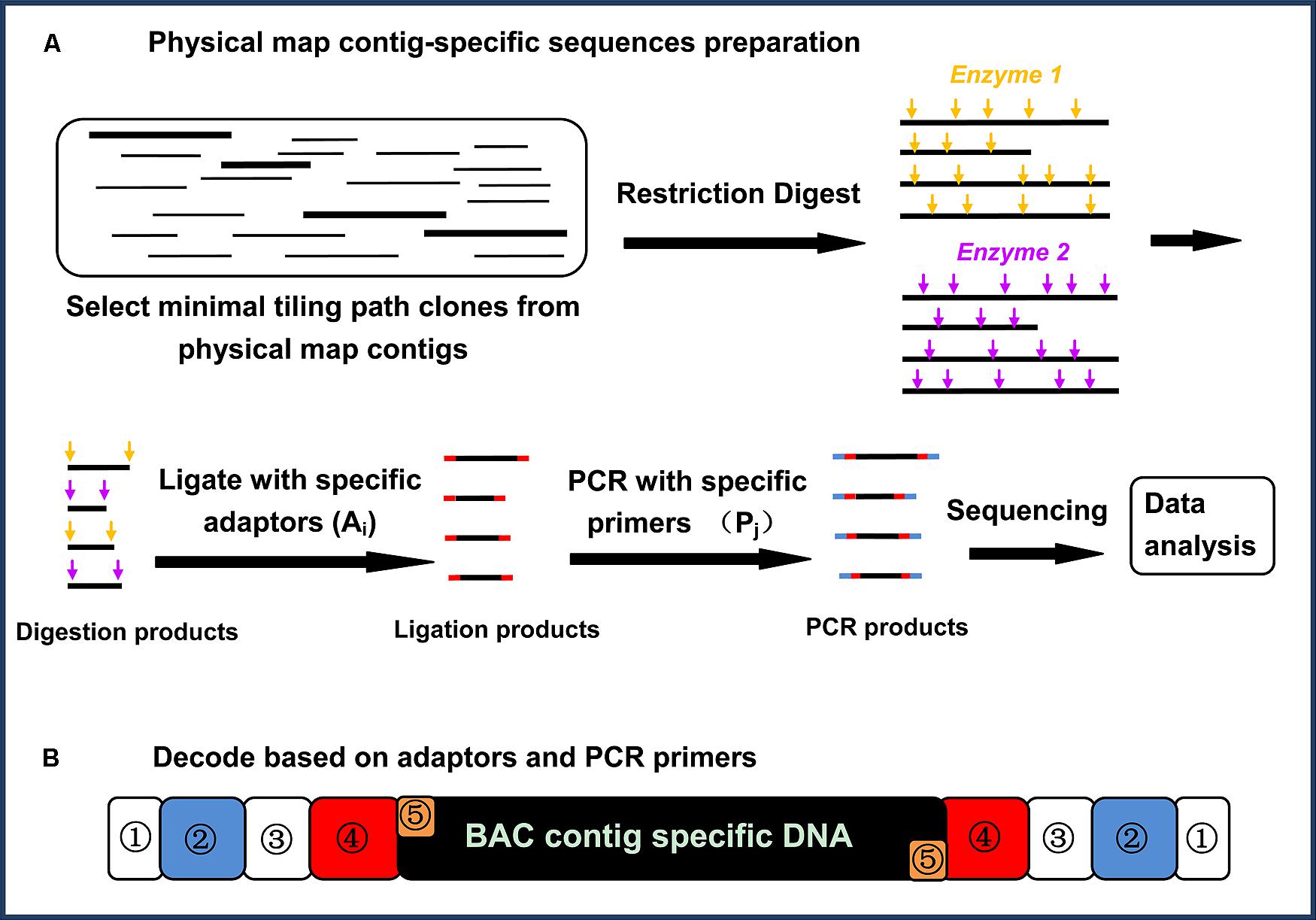
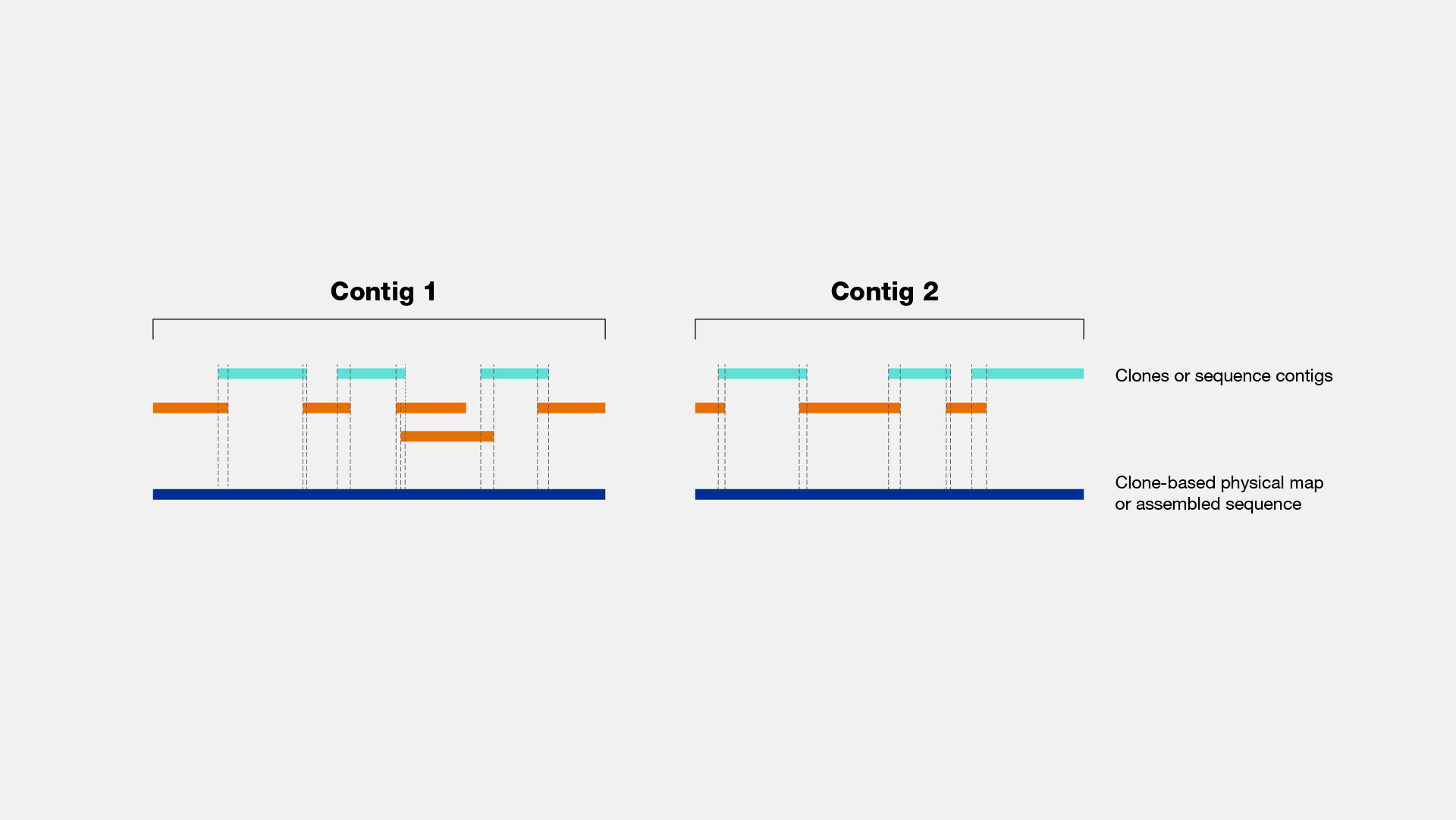
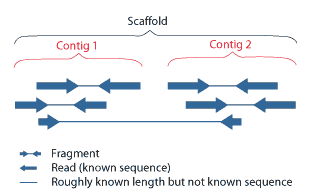
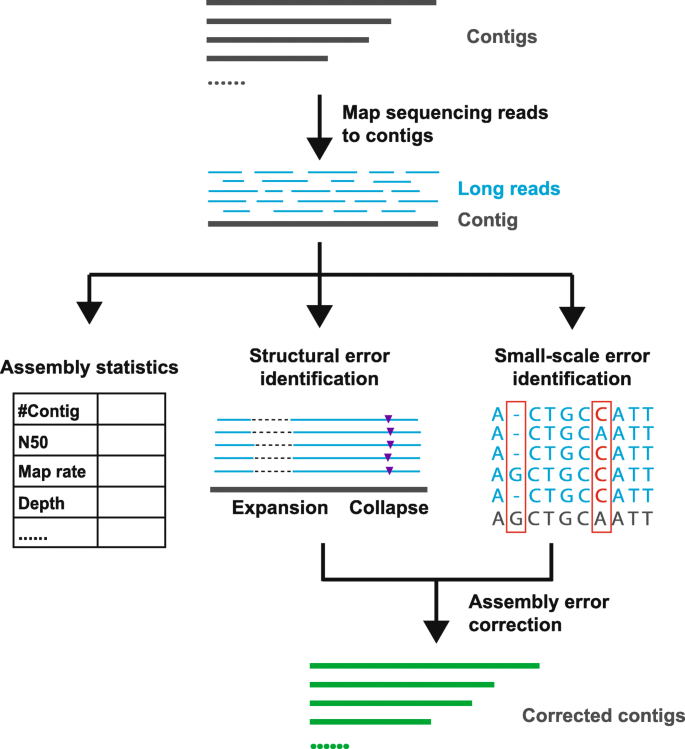
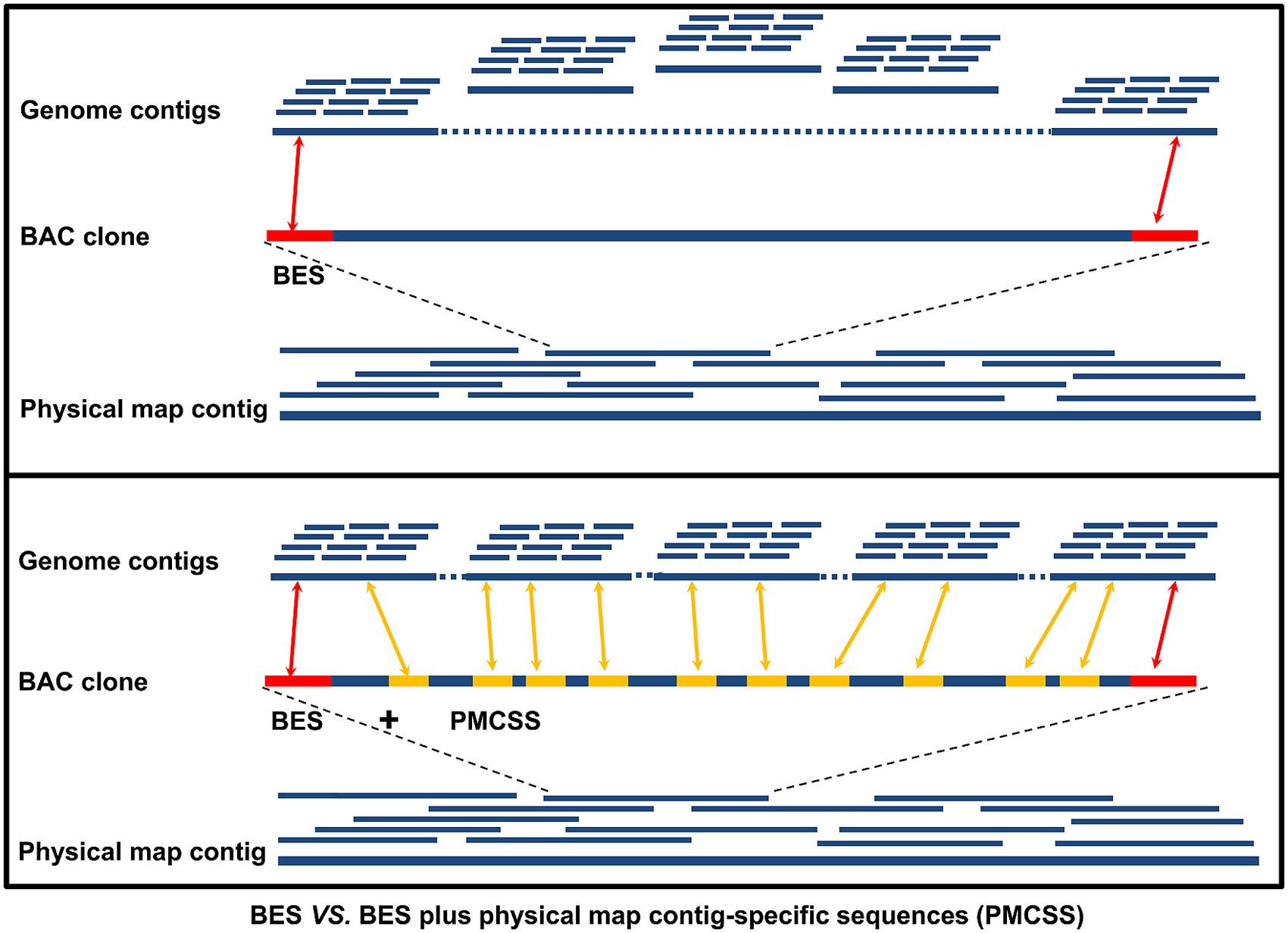
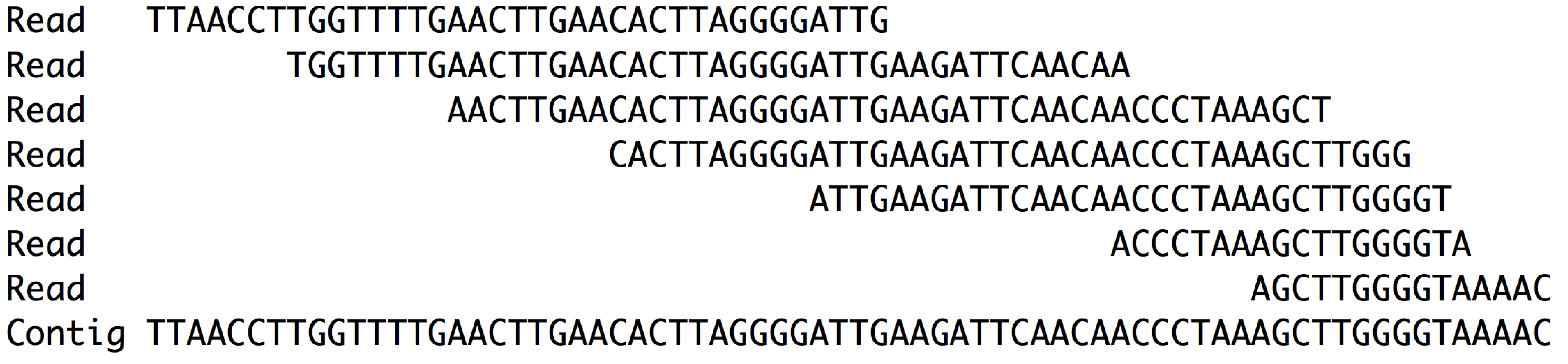Levels of clone and sequence coverage. A `®ngerprint clone contig' is... | Download Scientific Diagram

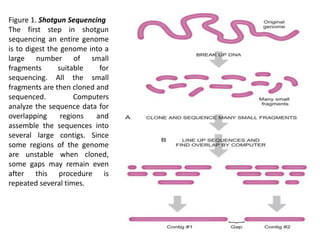
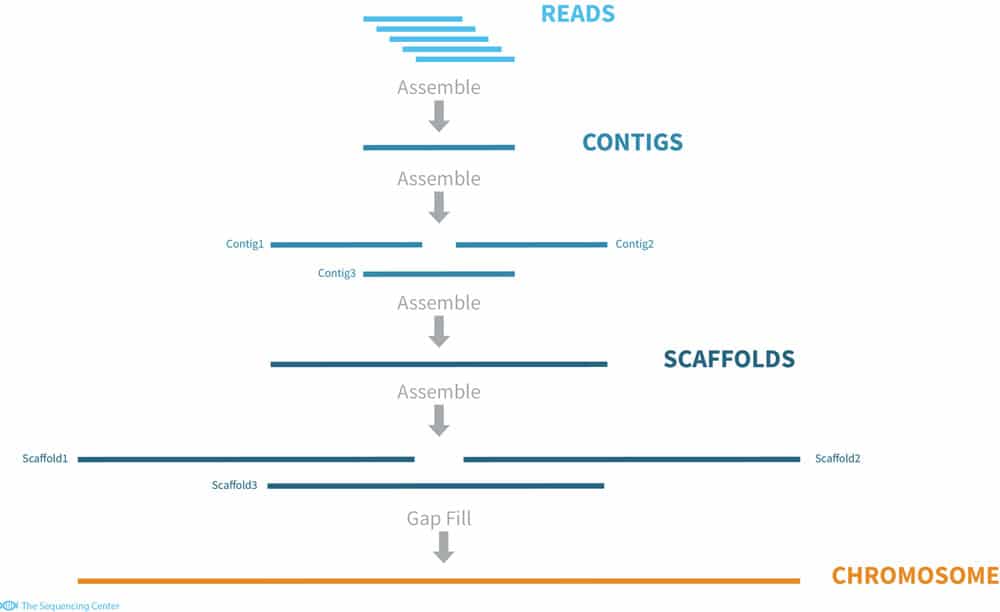
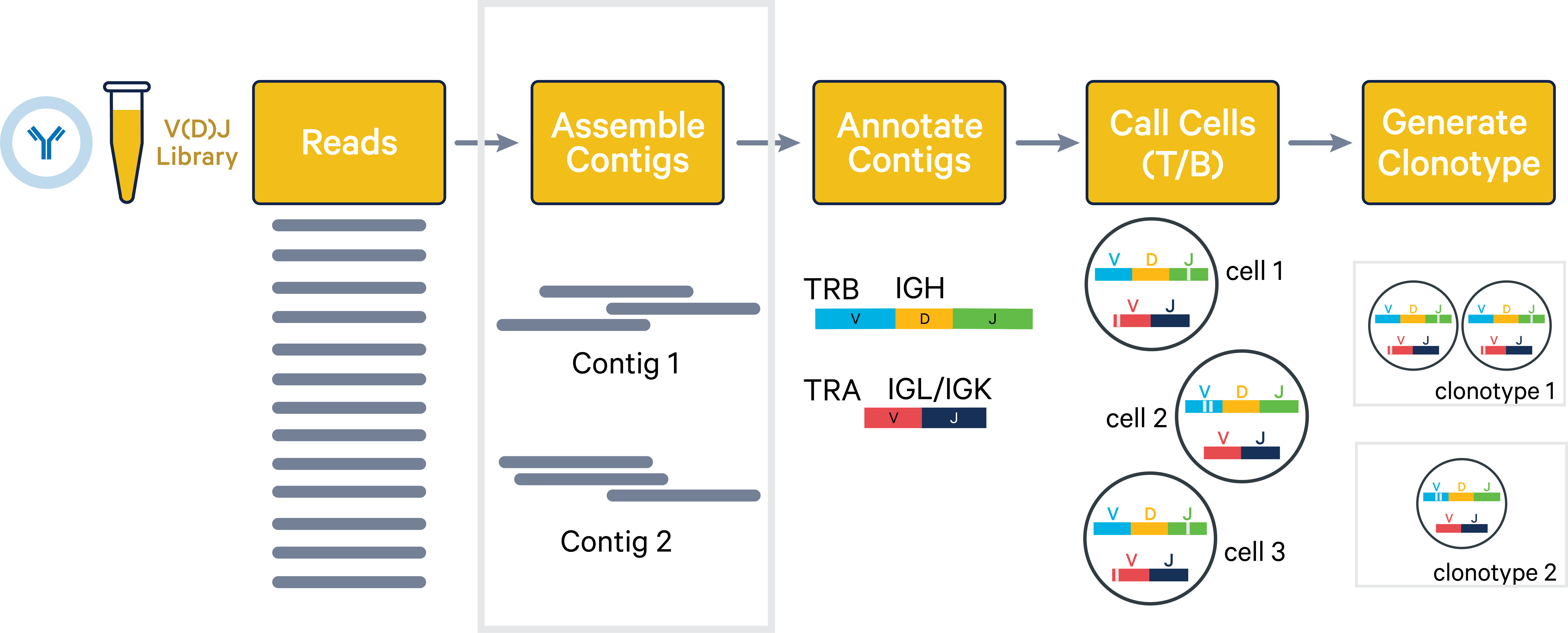
Whole Genome Sequencing Process of Creating Contigs from Fragment Reads | Download Scientific Diagram

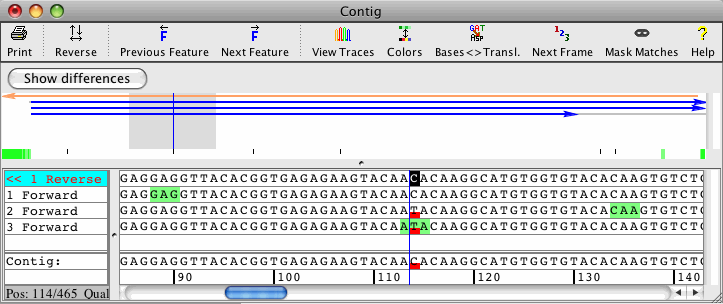
Clone Contig Sequencing Methods | Clone Contig Approach | Whole Genome Sequencing By Clone Contig | - YouTube
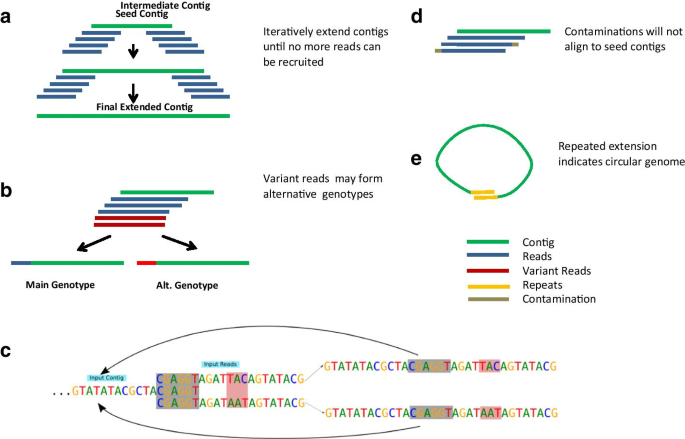
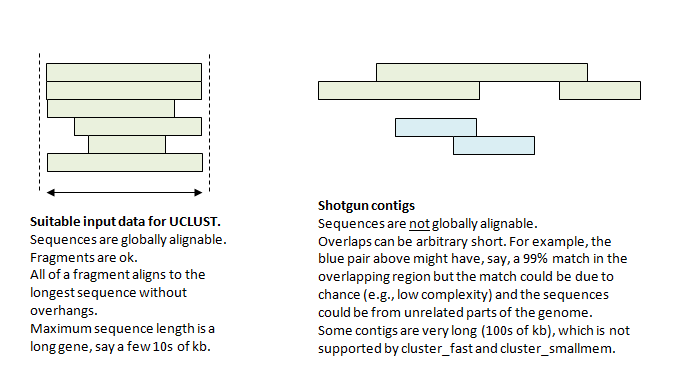
ContigExtender: a new approach to improving de novo sequence assembly for viral metagenomics data | BMC Bioinformatics | Full Text
Occupancy Modeling, Maximum Contig Size Probabilities and Designing Metagenomics Experiments | PLOS ONE

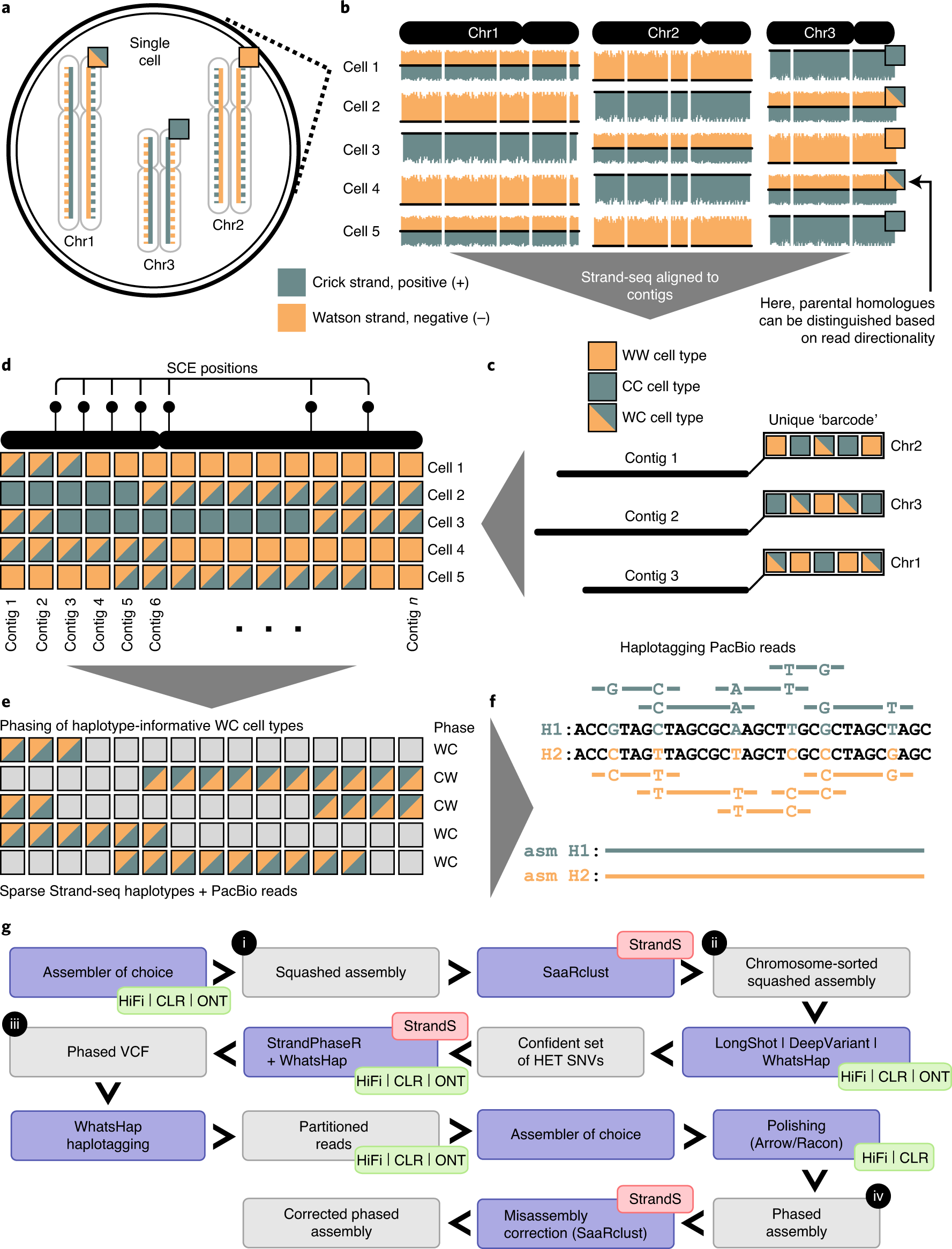
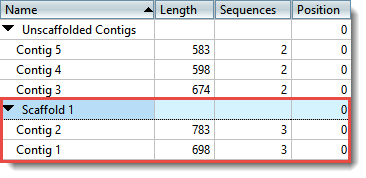
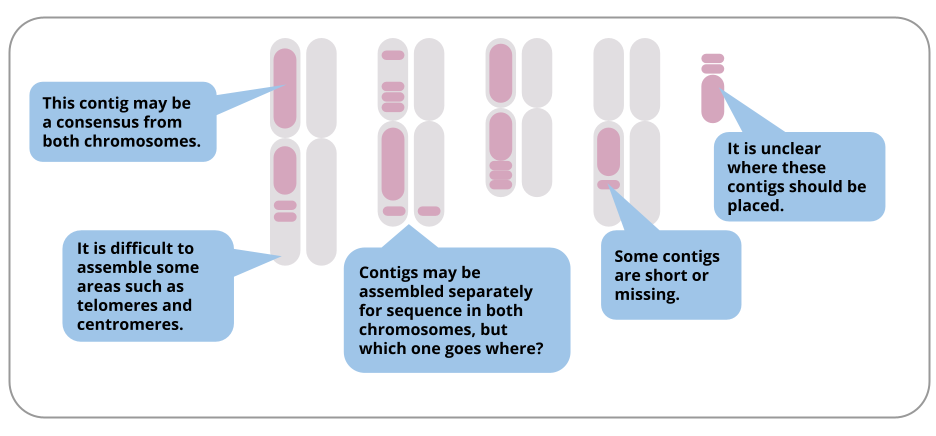
Types of assembled contigs and alignments to REF contigs. a A contig... | Download Scientific Diagram