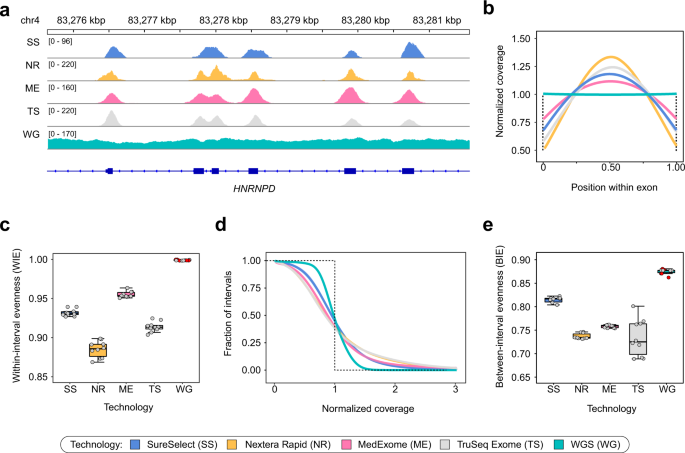
Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores | Genome Medicine | Full Text
GitHub - GenomicaMicrob/coverage_calculator: A simple script to calculate the coverage of a genome assembly

Mean mapped depth and coverage of diagnostic genomic regions according... | Download Scientific Diagram
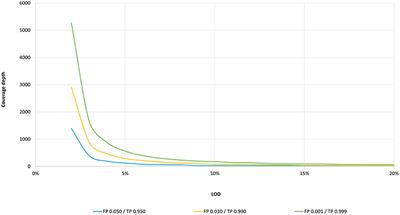
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics
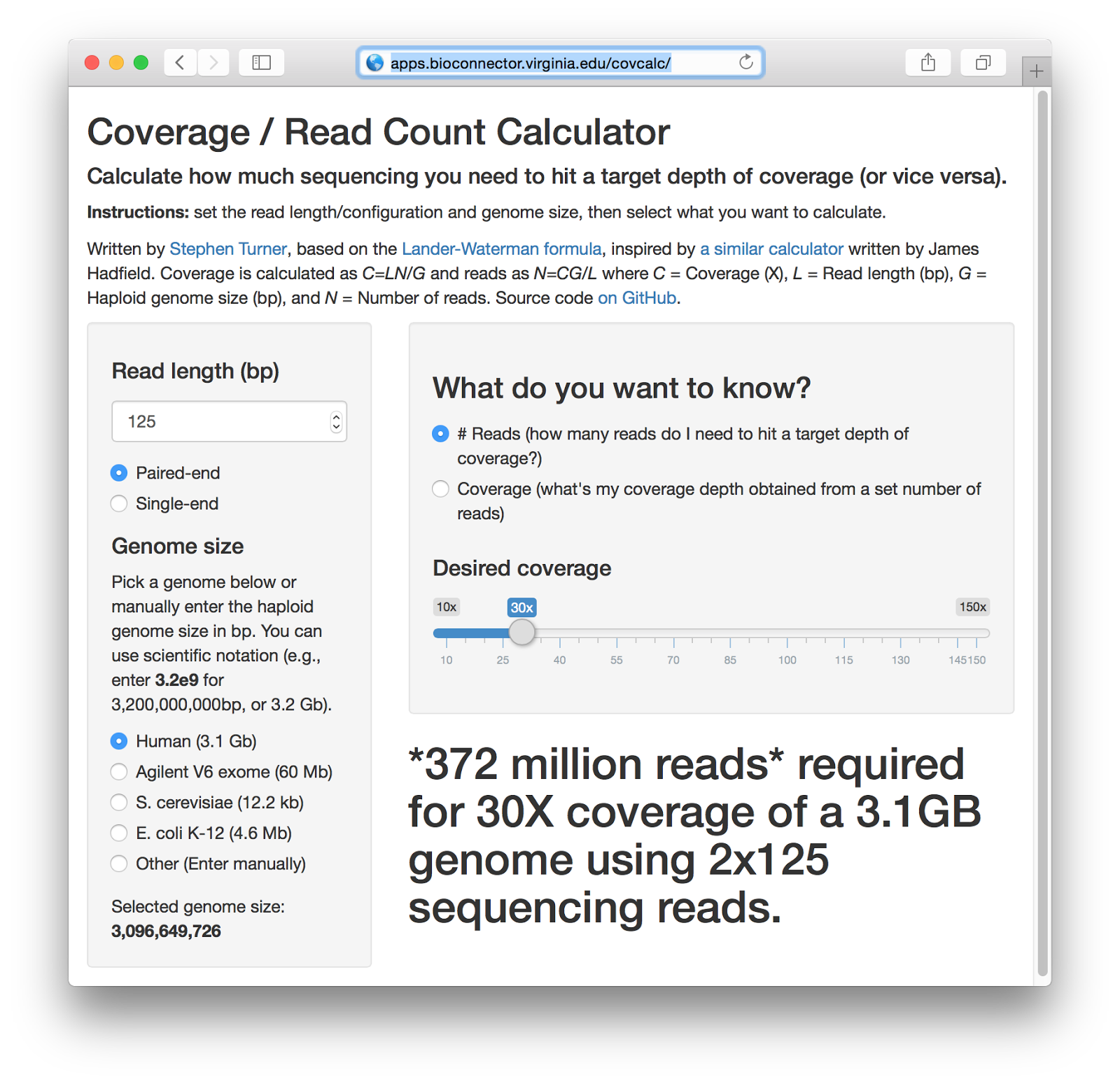
Getting Genetics Done: Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments

Recommendations for accurate genotyping of SARS-CoV-2 using amplicon-based sequencing of clinical samples - ScienceDirect

Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers

Sample depth of coverage. Histogram of the mean sequencing read depth... | Download Scientific Diagram