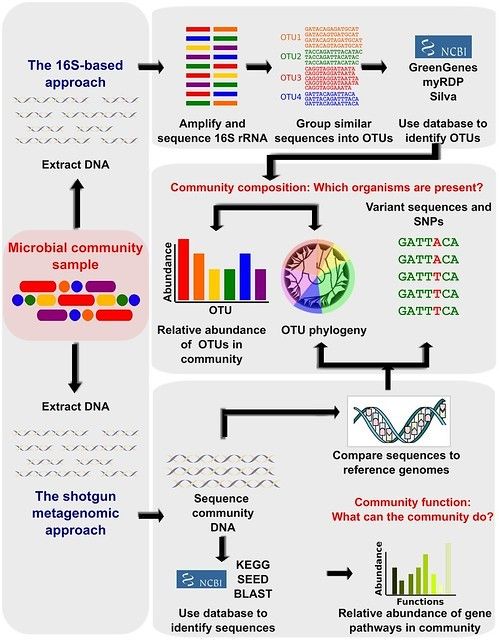
Strengths and Limitations of 16S rRNA Gene Amplicon Sequencing in Revealing Temporal Microbial Community Dynamics | PLOS ONE

Multicenter assessment of microbial community profiling using 16S rRNA gene sequencing and shotgun metagenomic sequencing - ScienceDirect
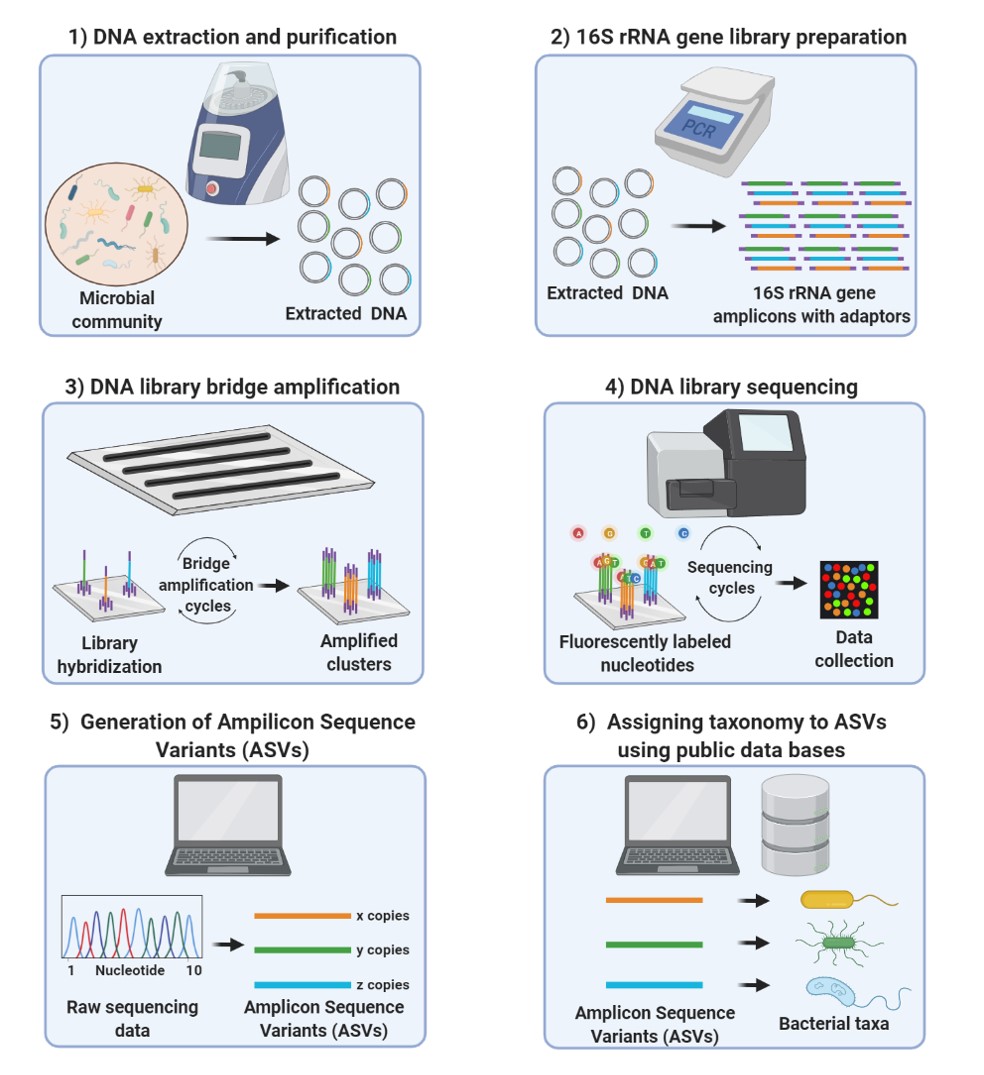
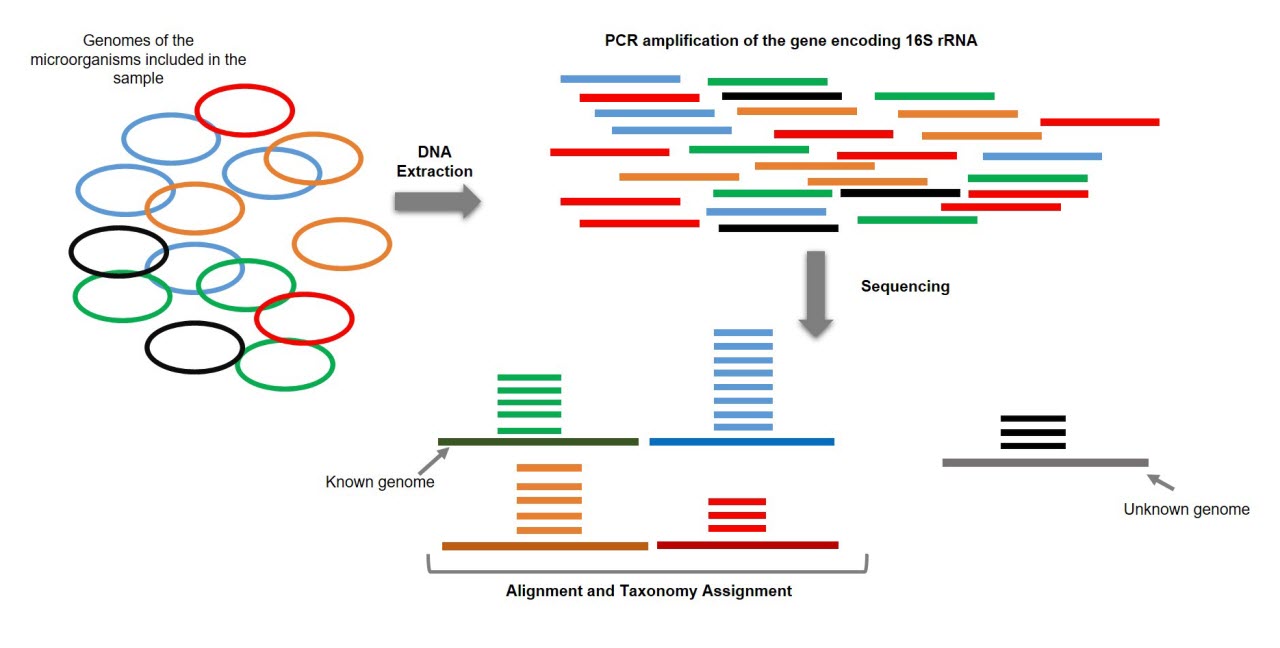
Demonstration of the workflow for the 16S rRNA and ITS amplicon sequencing. | Download Scientific Diagram
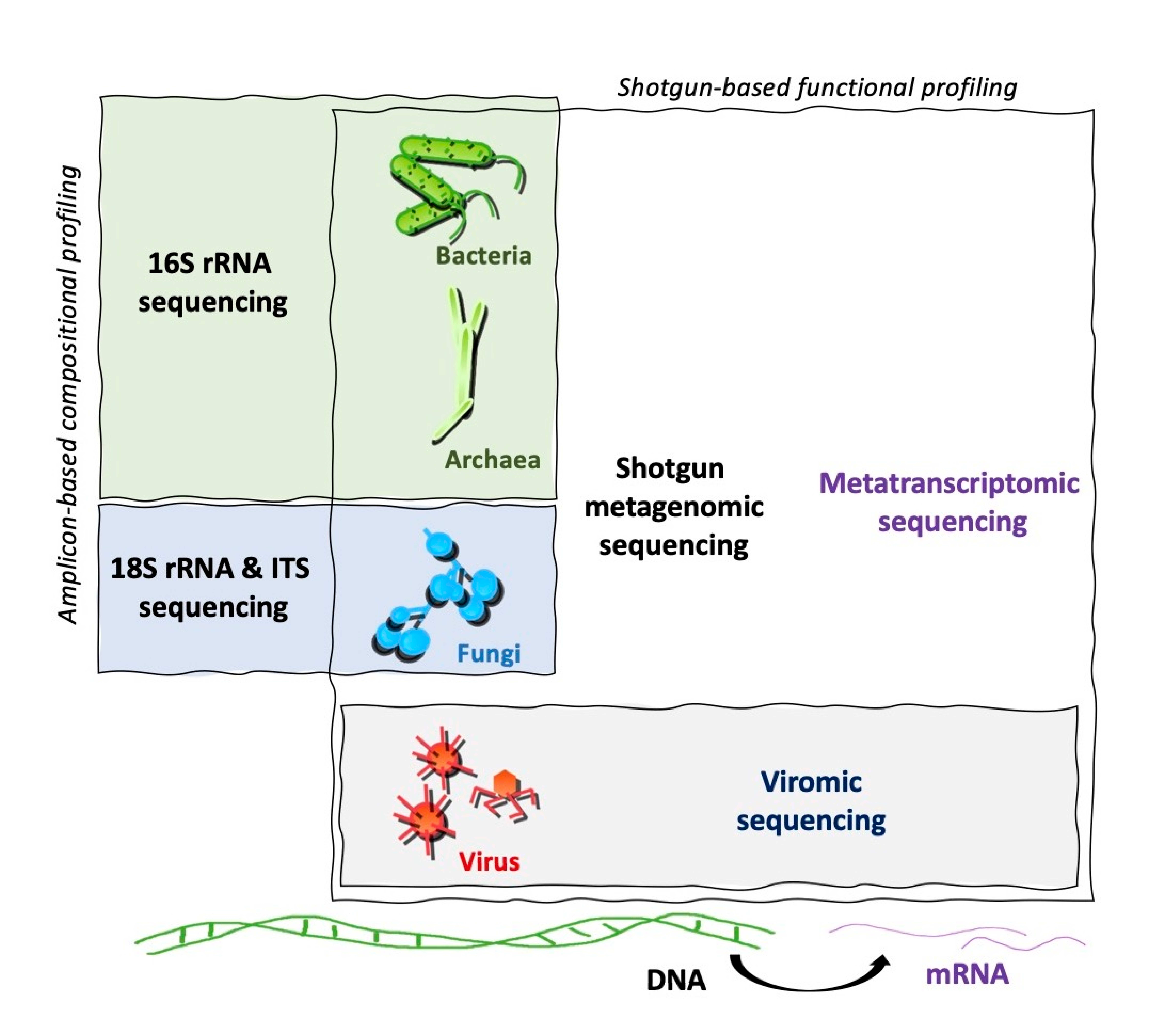
Biomolecules | Free Full-Text | An Introduction to Next Generation Sequencing Bioinformatic Analysis in Gut Microbiome Studies
![PDF] A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq | Semantic Scholar PDF] A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/c5433f6643987939f74806d2f91db31fe7eac03a/4-Figure1-1.png)
PDF] A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq | Semantic Scholar
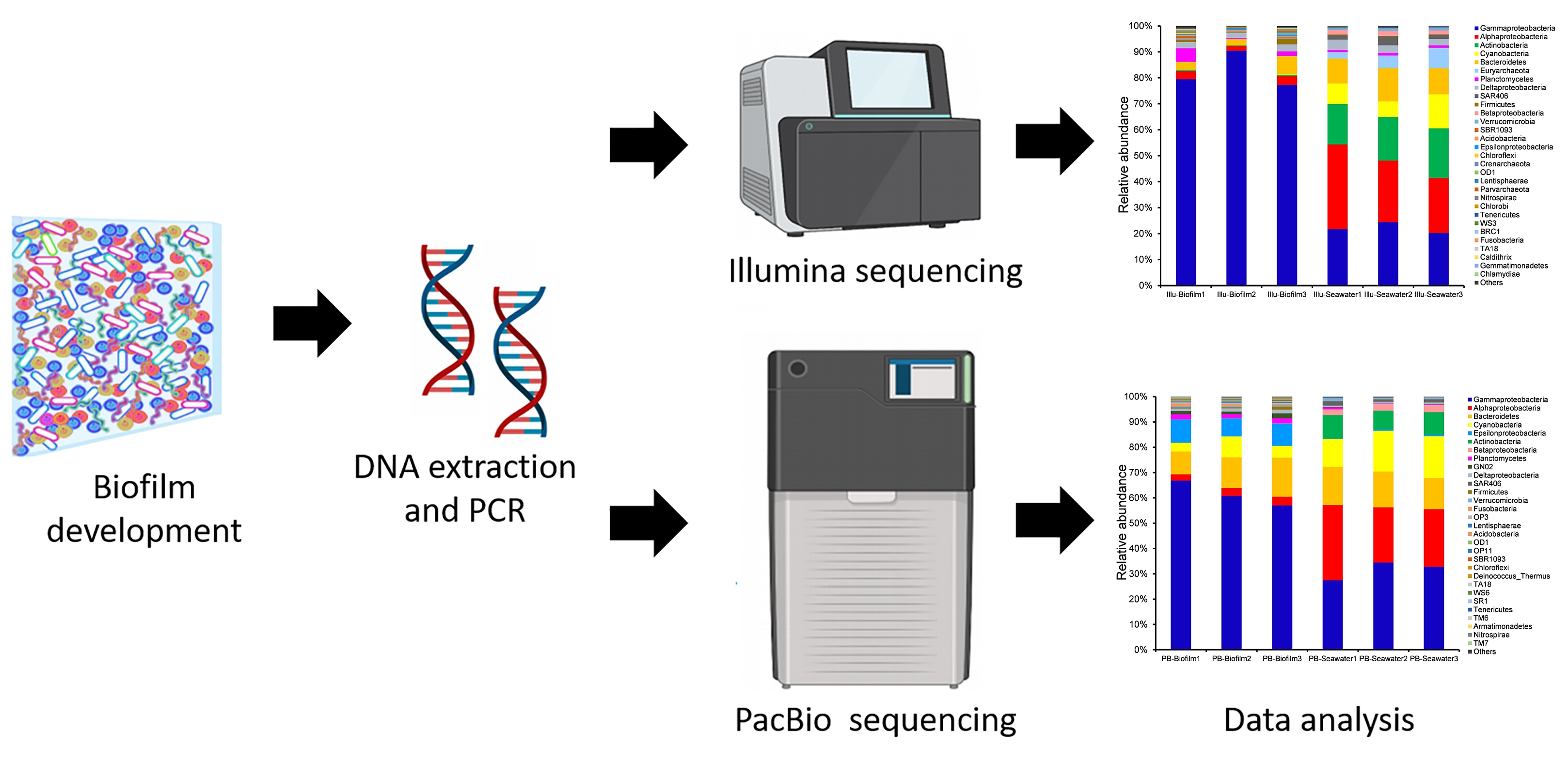
Genes | Free Full-Text | Microbial Richness of Marine Biofilms Revealed by Sequencing Full-Length 16S rRNA Genes
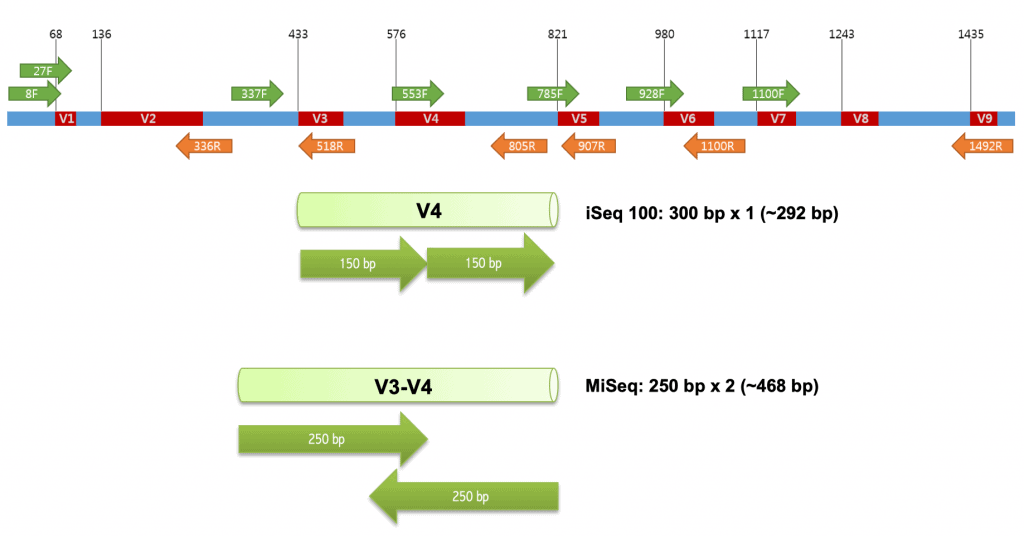
Highly multiplexed gene amplicon sequencing | Joint Microbiome Facility of the Medical University of Vienna and the University of Vienna



![De novo species identification using 16S rRNA gene nanopore sequencing [PeerJ] De novo species identification using 16S rRNA gene nanopore sequencing [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2020/10029/1/fig-1-full.png)