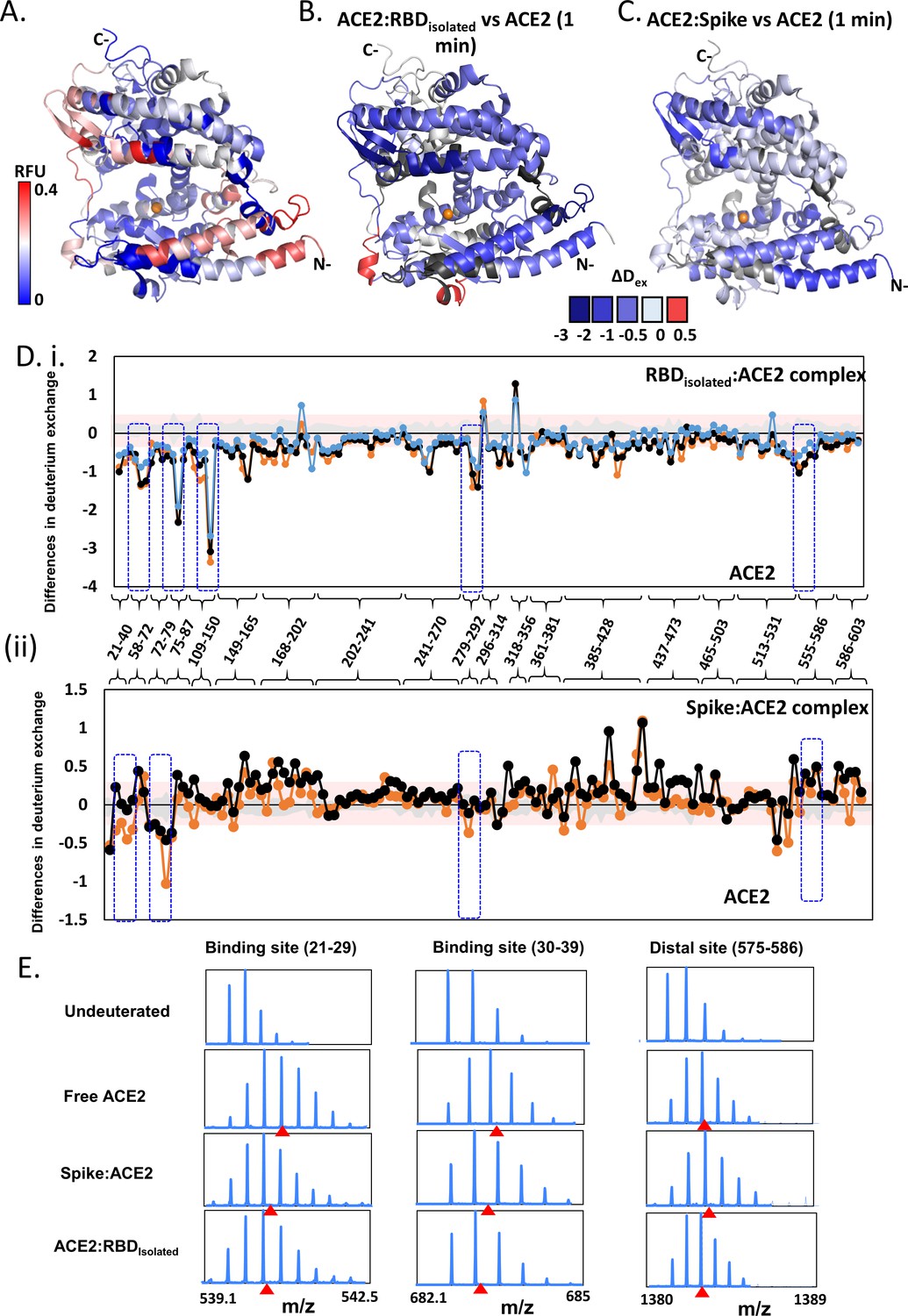
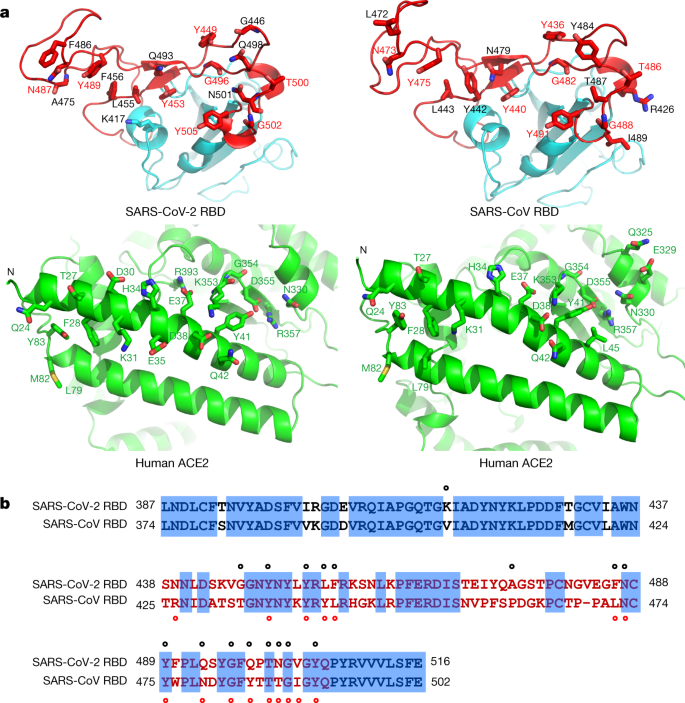
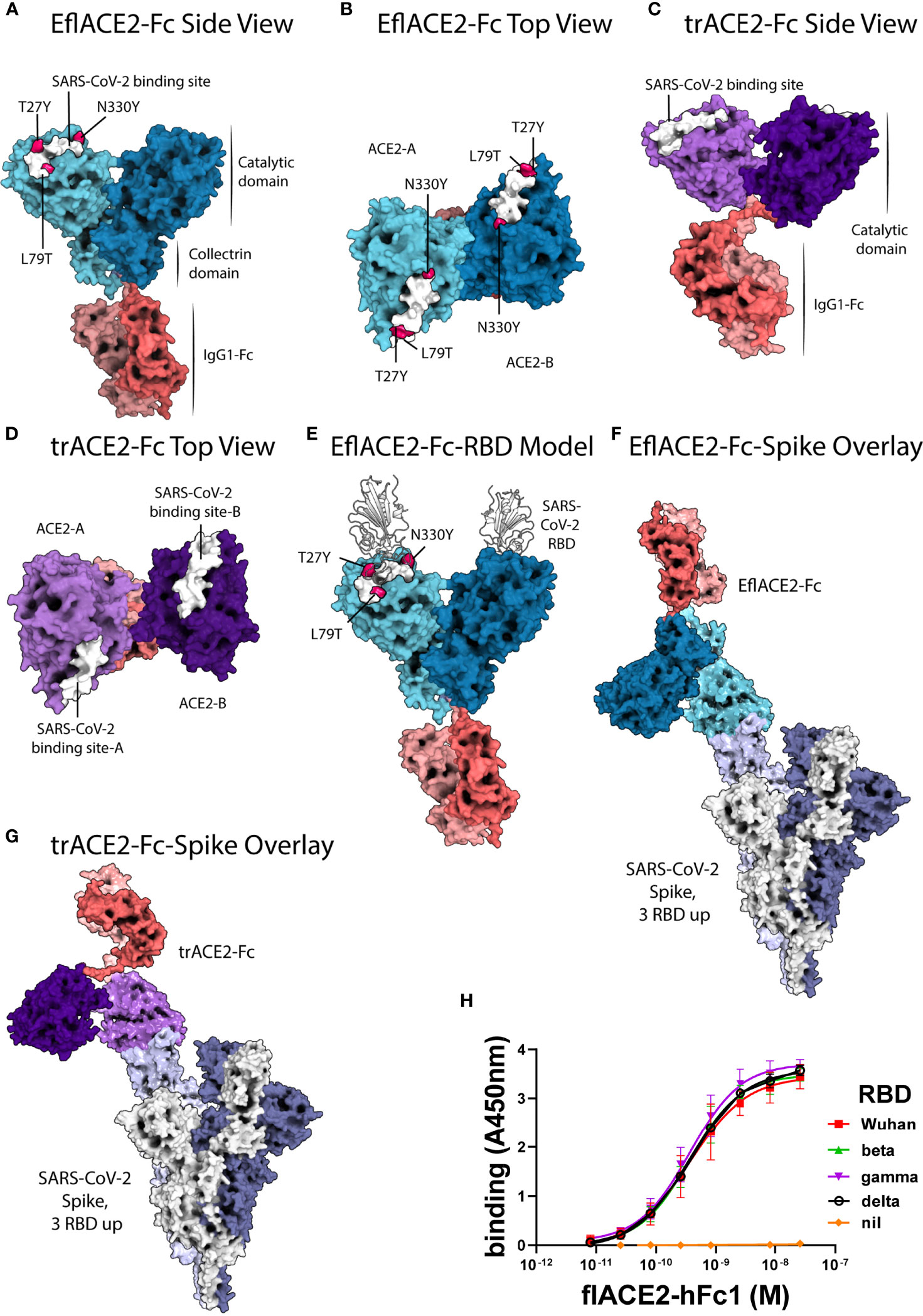
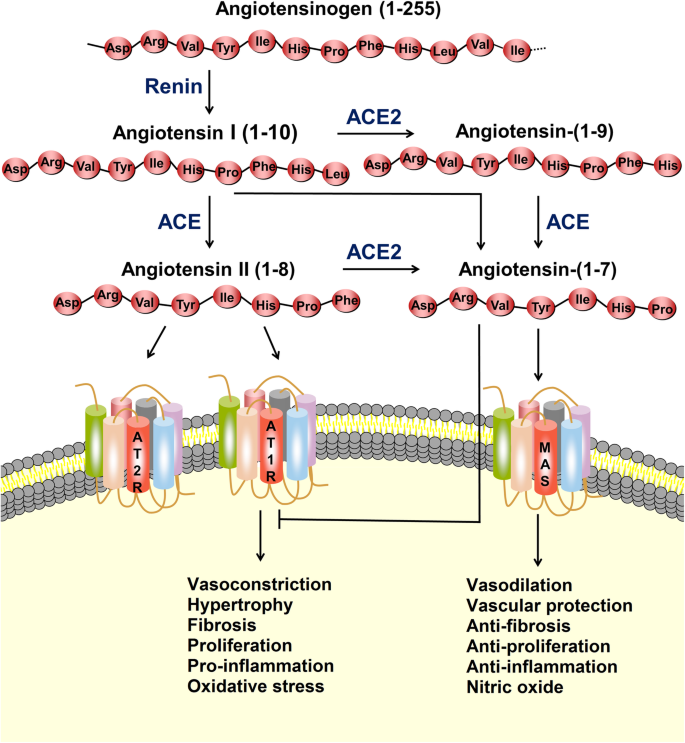
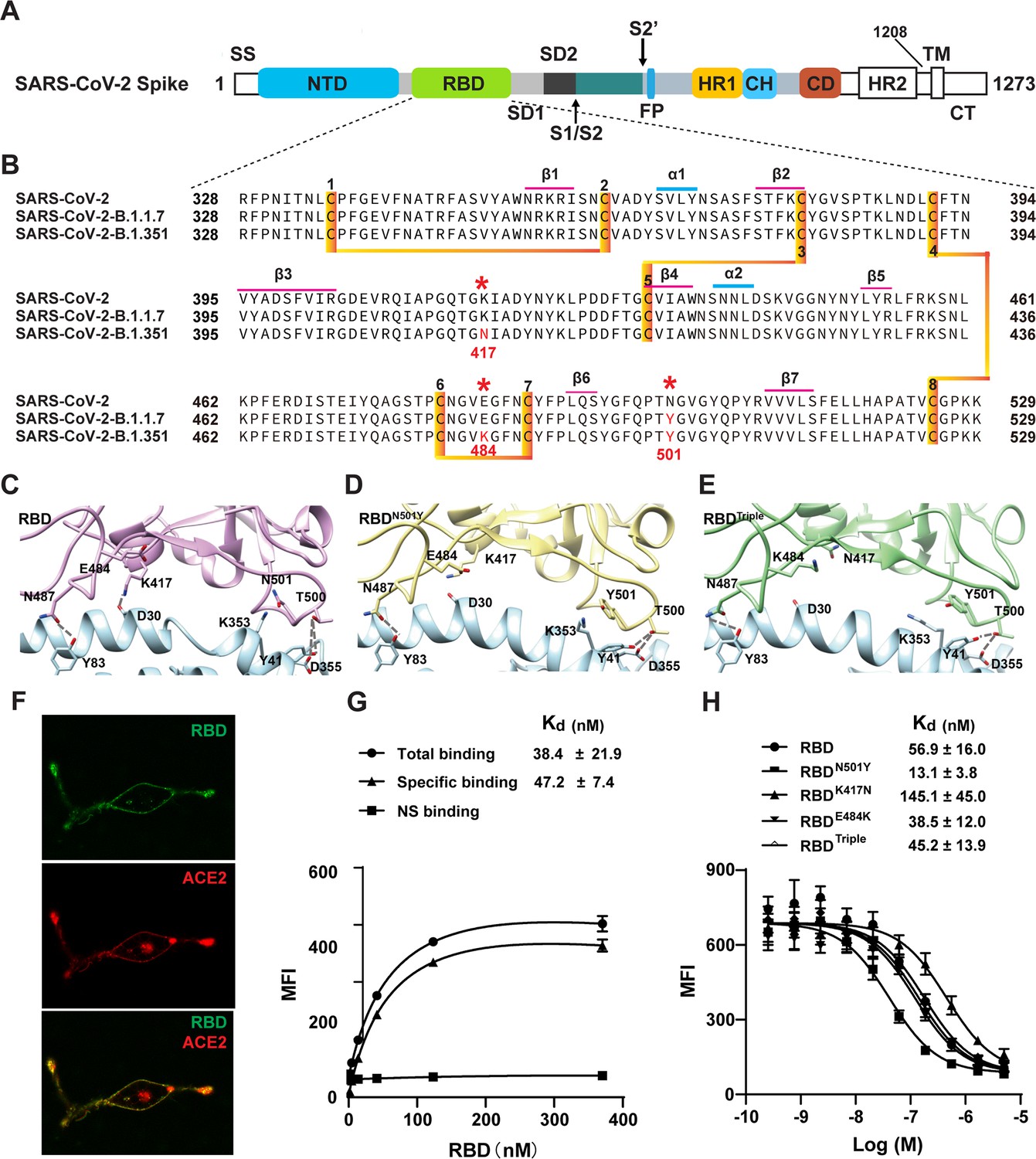
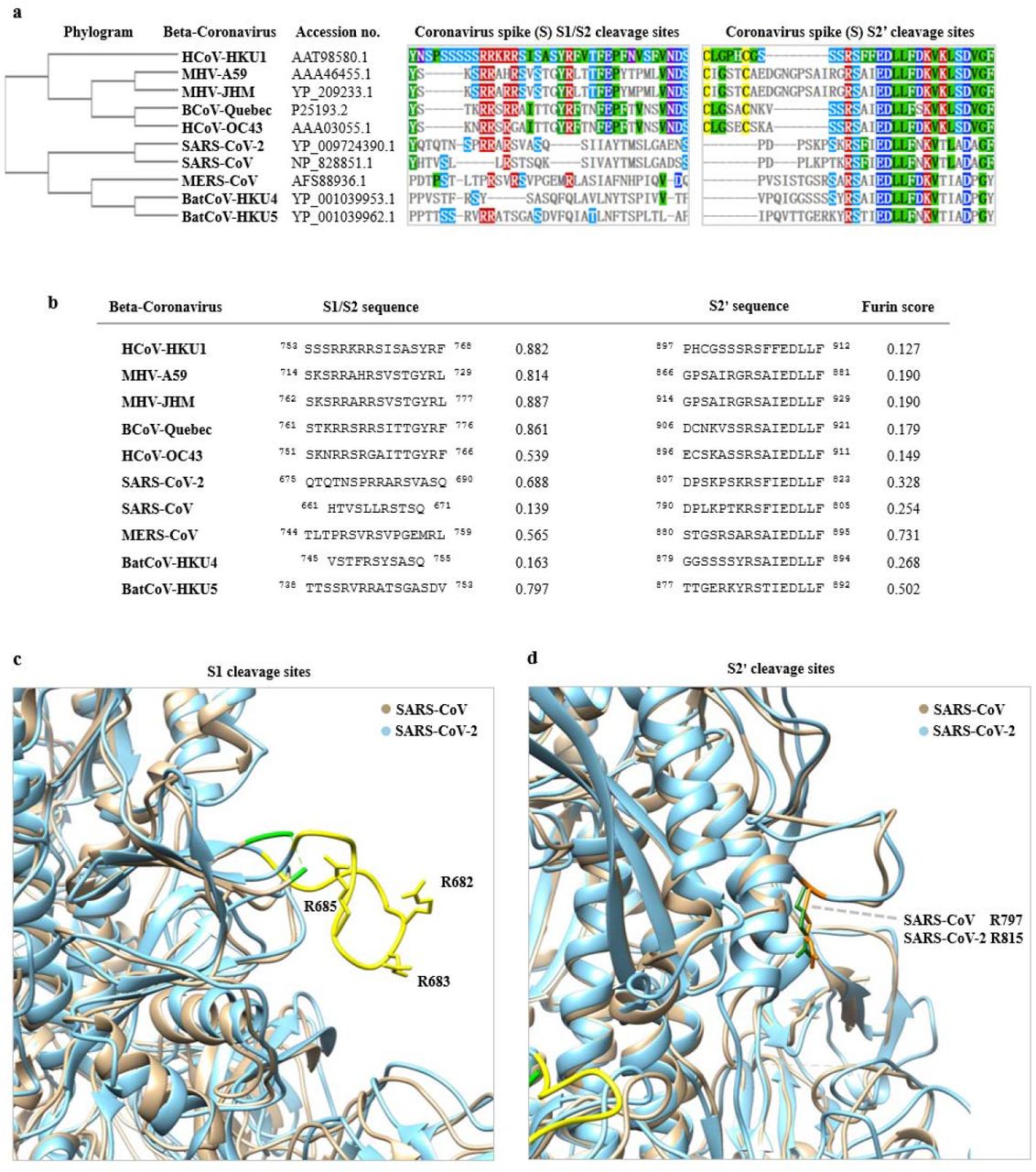
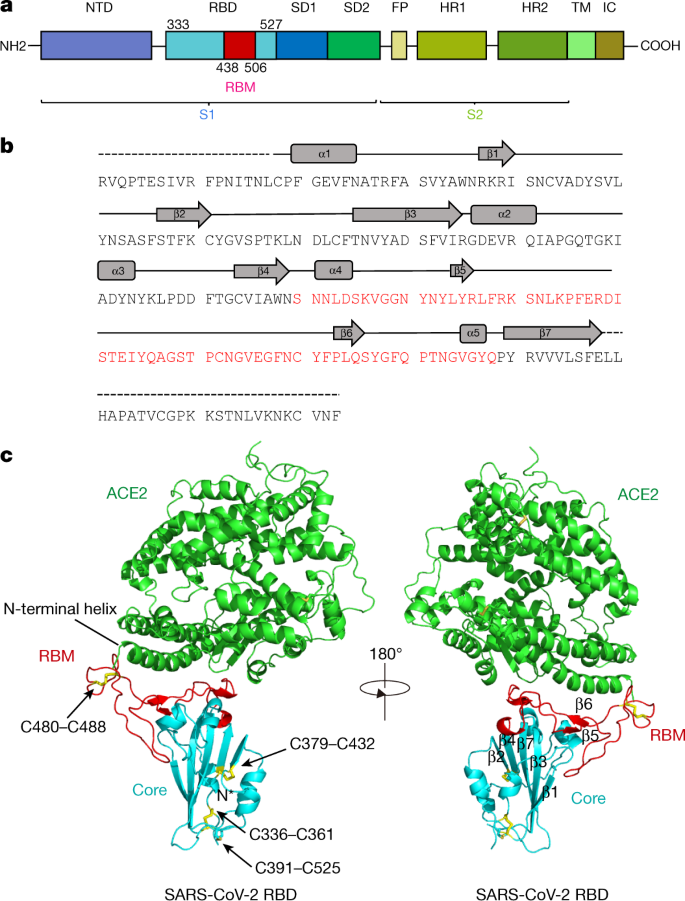
All-Atom Simulations of Human ACE2-Spike Protein RBD Complexes for SARS-CoV-2 and Some of its Variants: Nature of Interactions and Free Energy Diagrams for Dissociation of the Protein Complexes | The Journal of

SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex | Science

Critical Interactions Between the SARS-CoV-2 Spike Glycoprotein and the Human ACE2 Receptor | The Journal of Physical Chemistry B
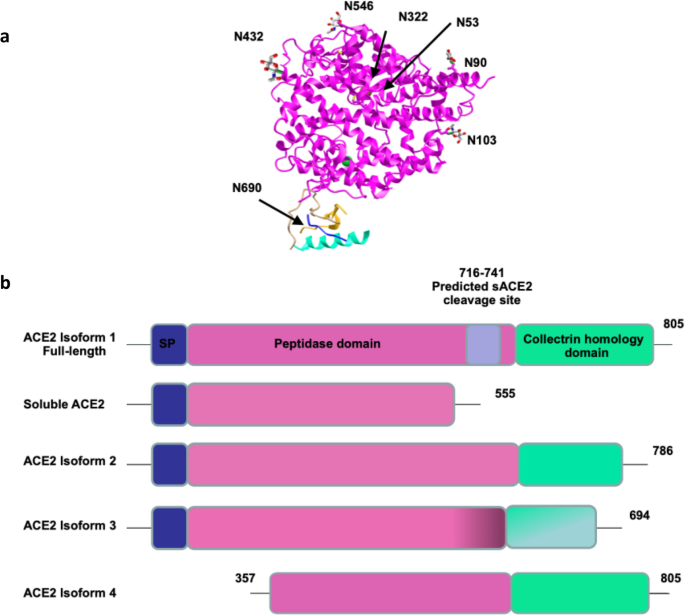
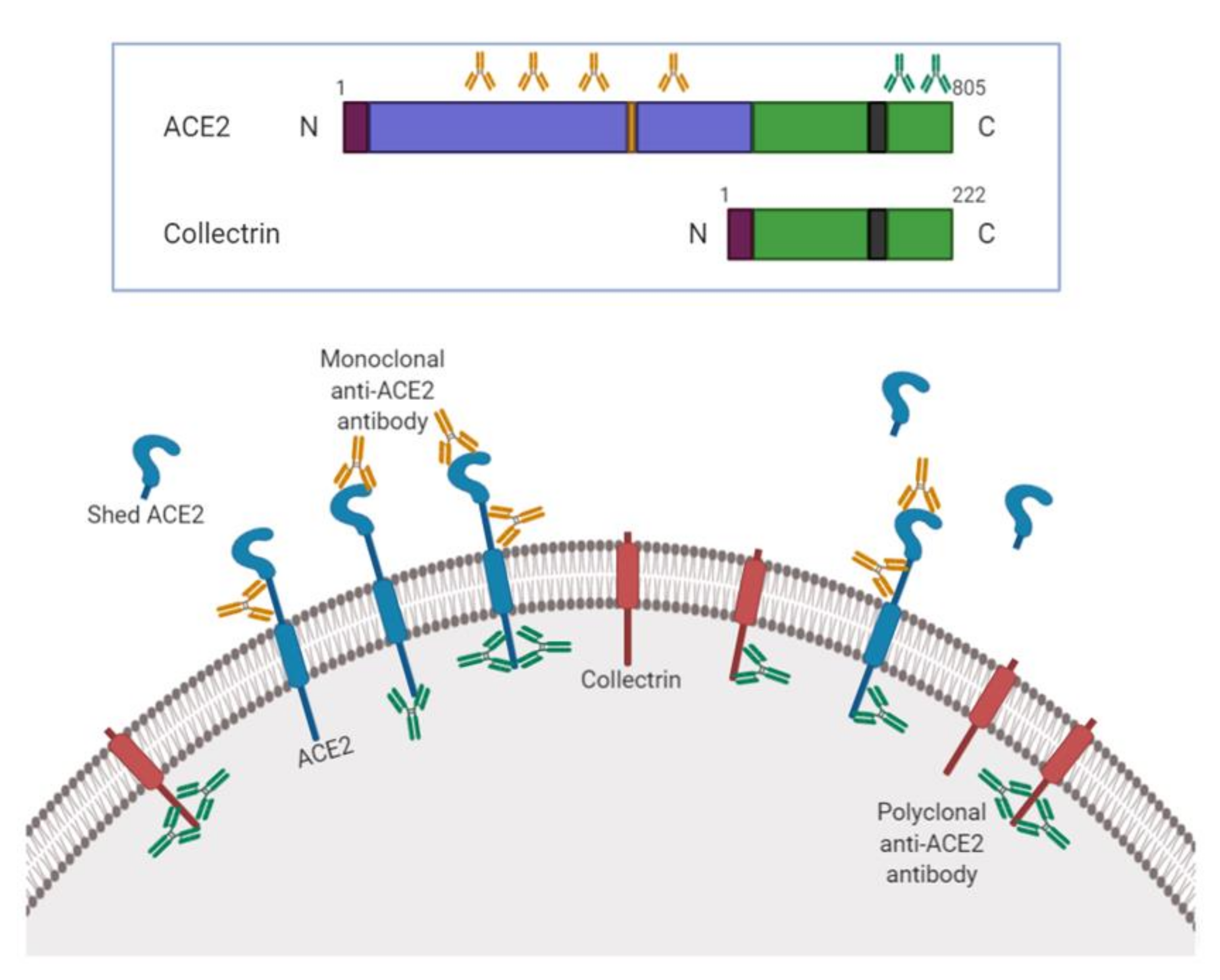
ACE2 Nascence, trafficking, and SARS-CoV-2 pathogenesis: the saga continues | Human Genomics | Full Text

Does SARS-CoV-2 Bind to Human ACE2 More Strongly Than Does SARS-CoV? | The Journal of Physical Chemistry B
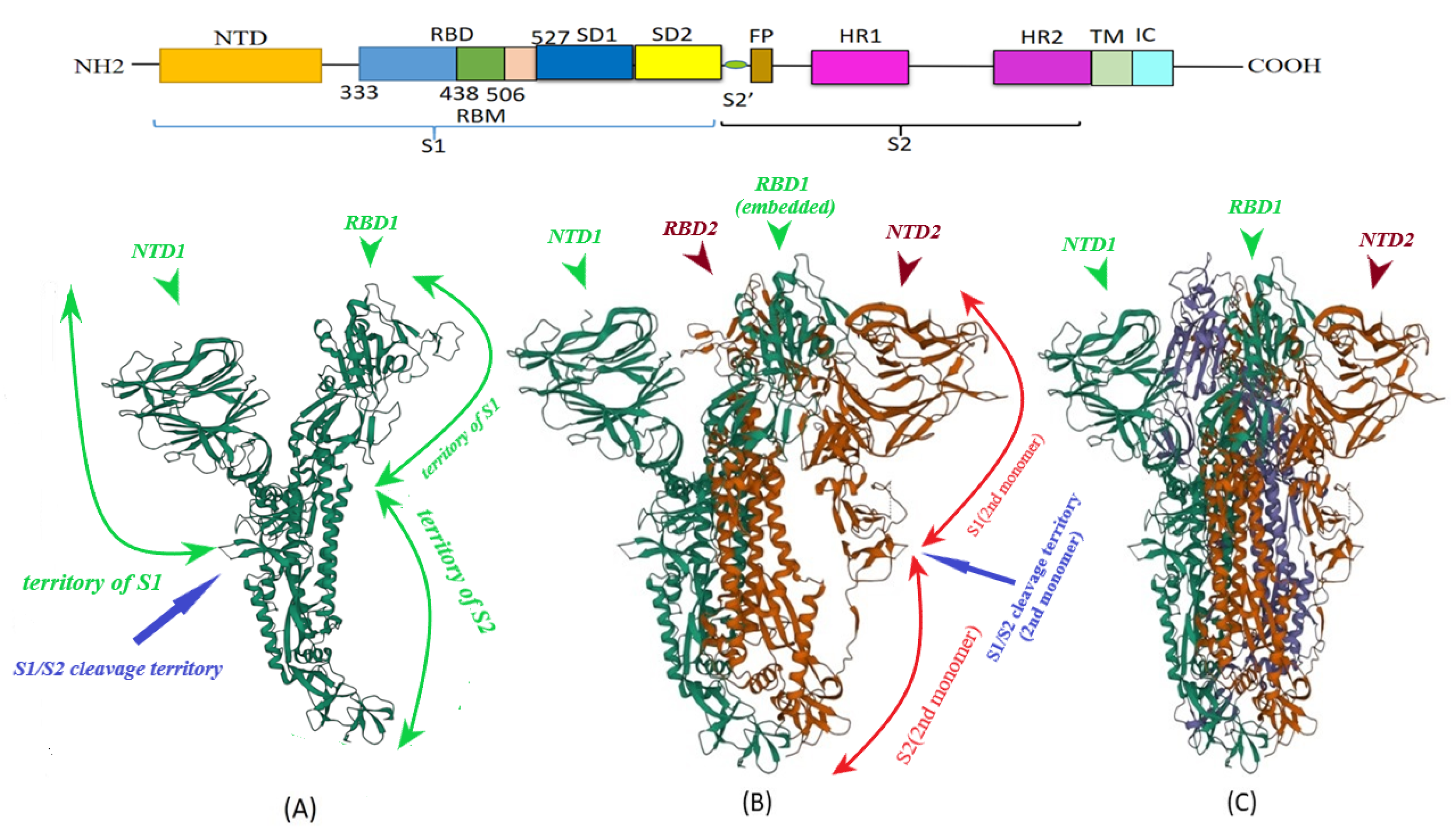
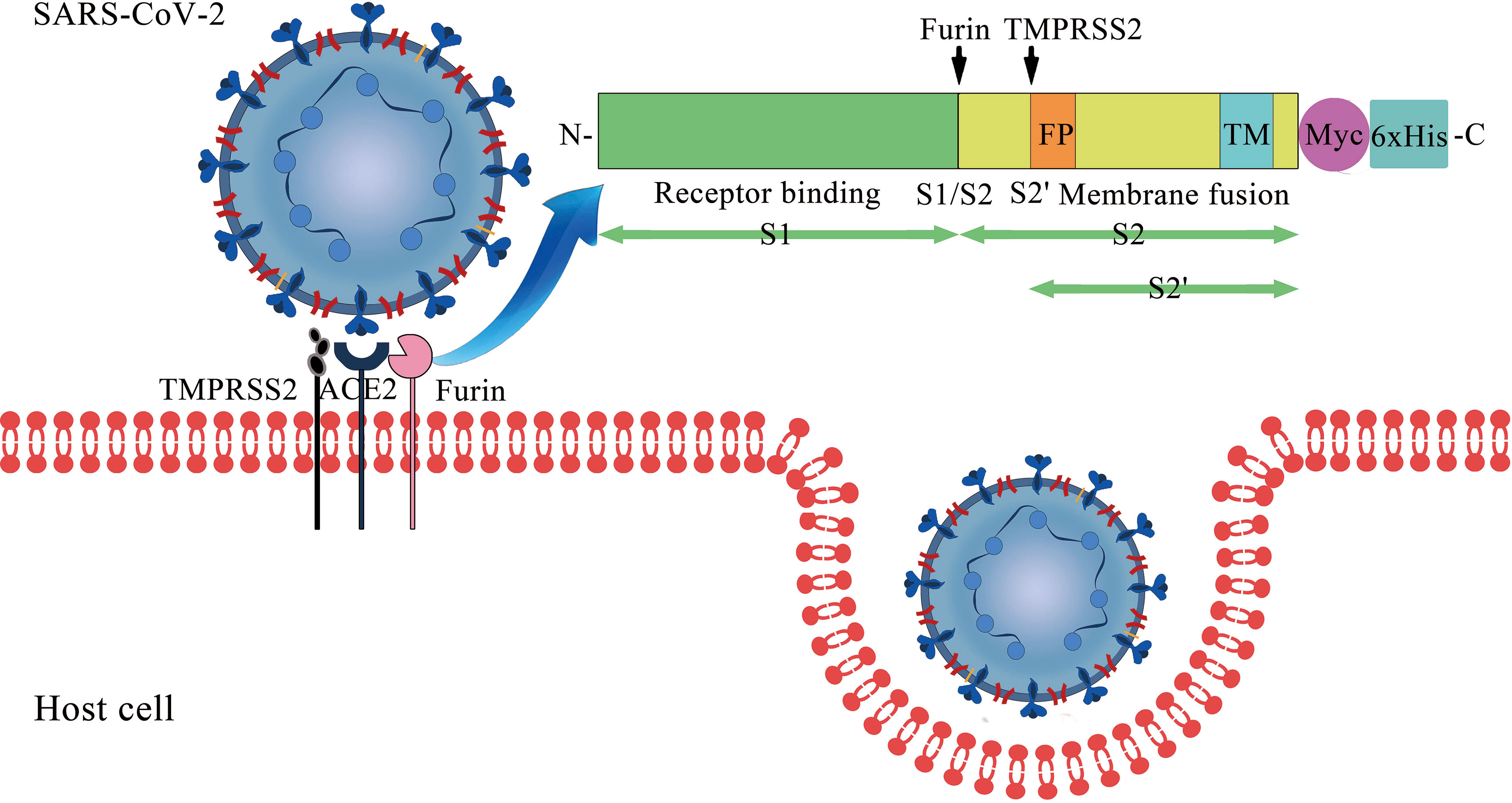
Vaccines | Free Full-Text | S Protein, ACE2 and Host Cell Proteases in SARS-CoV-2 Cell Entry and Infectivity; Is Soluble ACE2 a Two Blade Sword? A Narrative Review
Frontiers | The Impact of ACE2 Polymorphisms on COVID-19 Disease: Susceptibility, Severity, and Therapy
Biology | Free Full-Text | ACE2 Protein Landscape in the Head and Neck Region: The Conundrum of SARS-CoV-2 Infection

In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin | Scientific Reports

Structural models of human ACE2 variants with SARS-CoV-2 Spike protein for structure-based drug design | Scientific Data

Predicting Mutational Effects on Receptor Binding of the Spike Protein of SARS-CoV-2 Variants | Journal of the American Chemical Society

Computational biophysical characterization of the SARS-CoV-2 spike protein binding with the ACE2 receptor and implications for infectivity - ScienceDirect