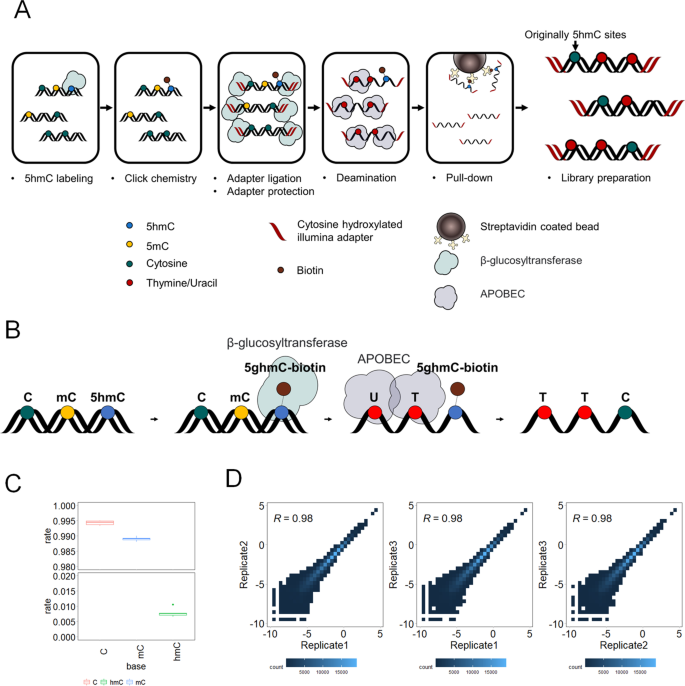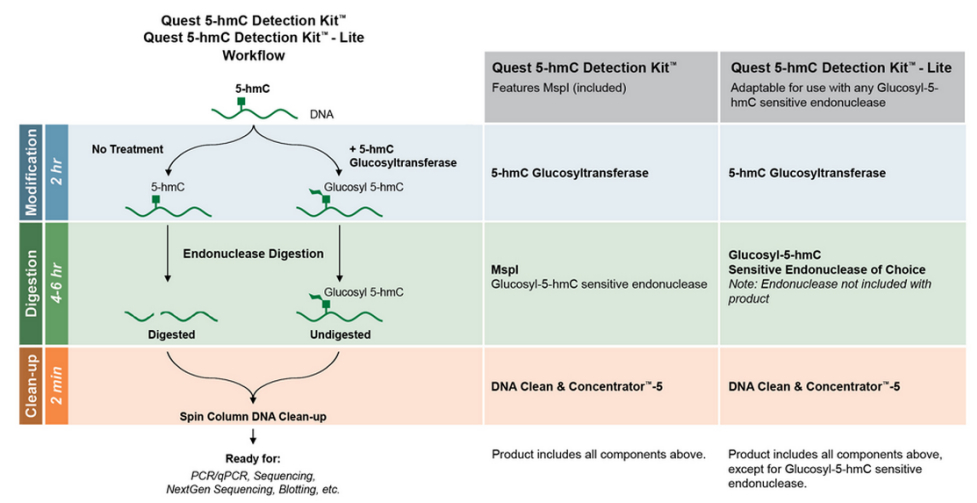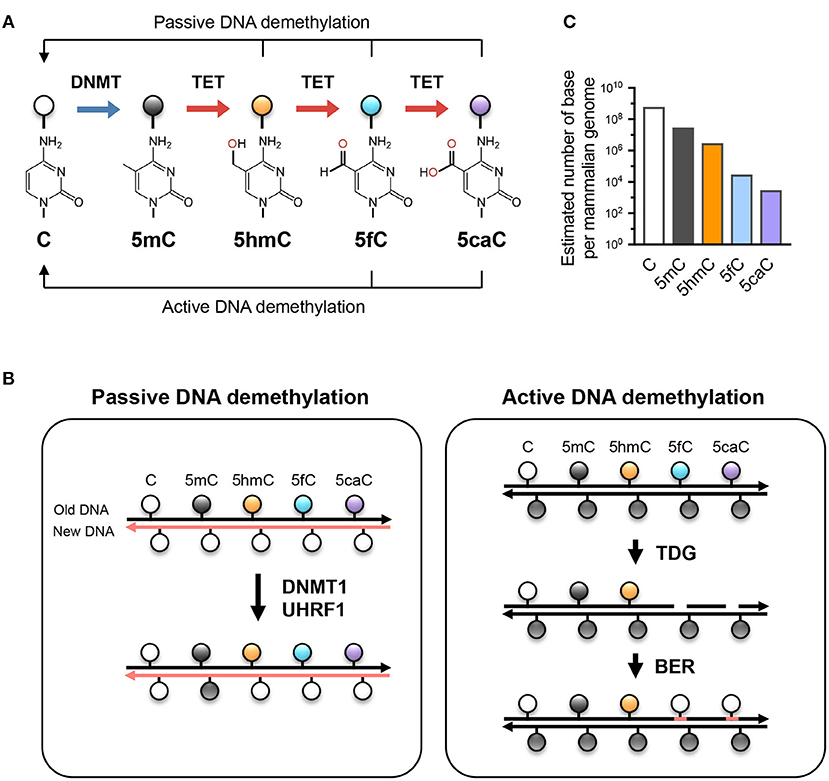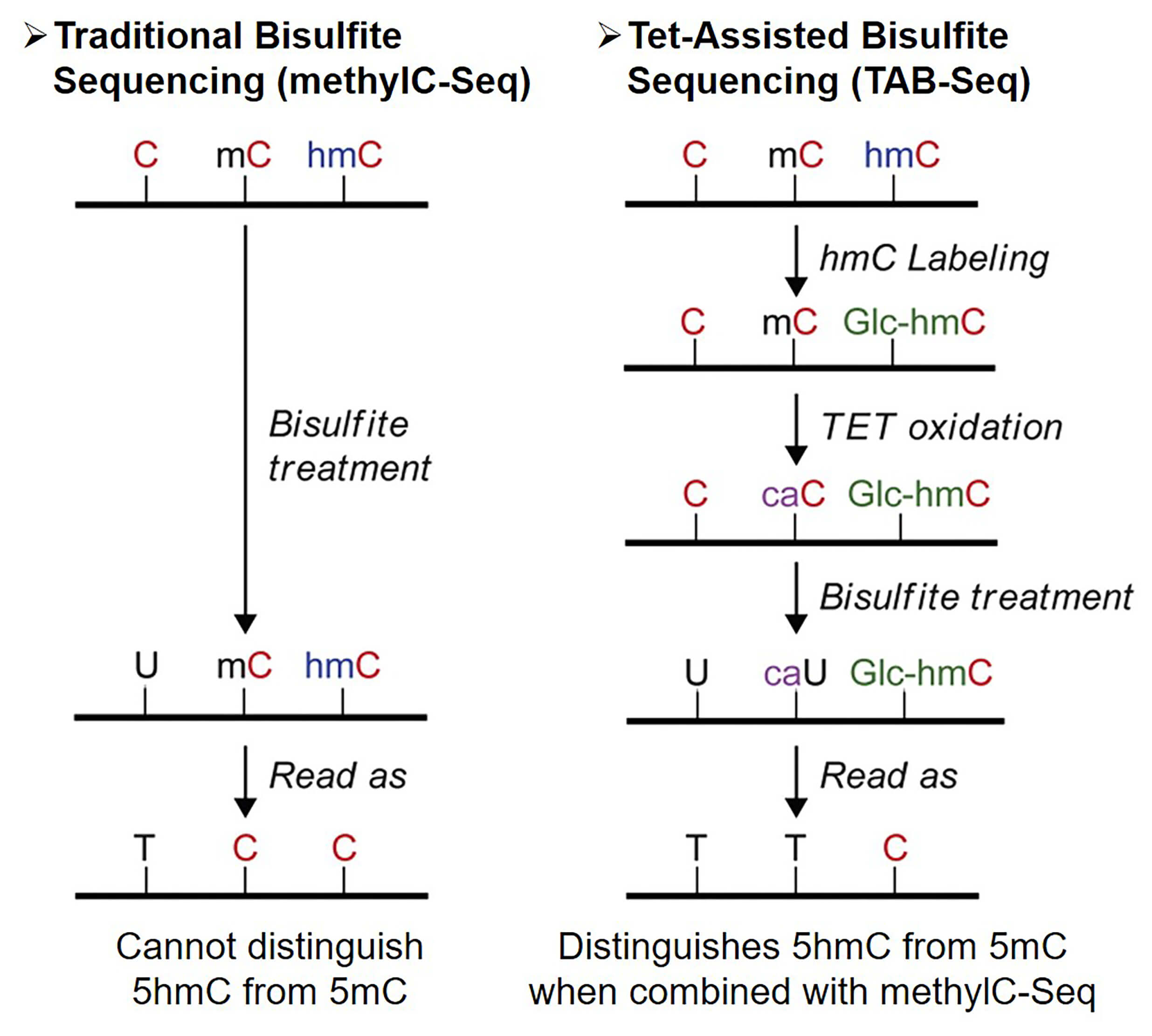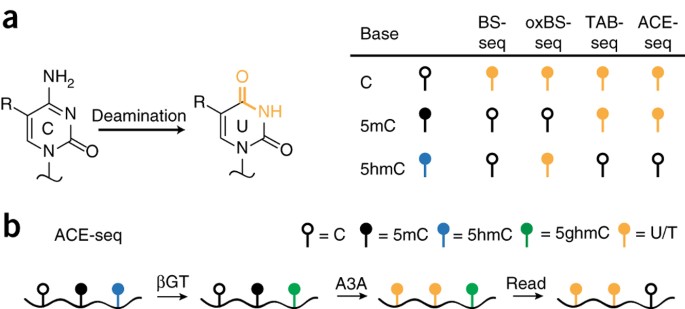
Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase | Nature Biotechnology
Frontiers | Selective Chemical Labeling and Sequencing of 5-Hydroxymethylcytosine in DNA at Single-Base Resolution

Bisulfite-Free, Nanoscale Analysis of 5-Hydroxymethylcytosine at Single Base Resolution | Journal of the American Chemical Society

Jump-seq: Genome-Wide Capture and Amplification of 5-Hydroxymethylcytosine Sites | Journal of the American Chemical Society
Precise genomic mapping of 5-hydroxymethylcytosine via covalent tether-directed sequencing | PLOS Biology

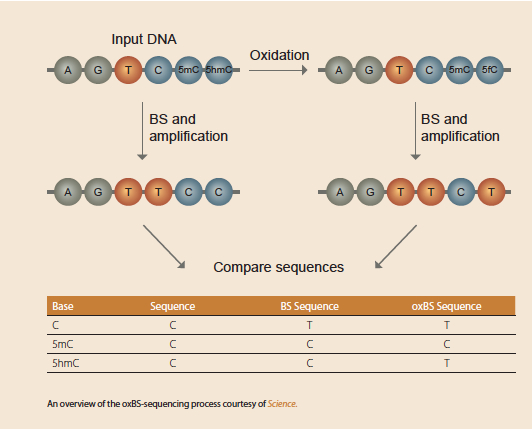
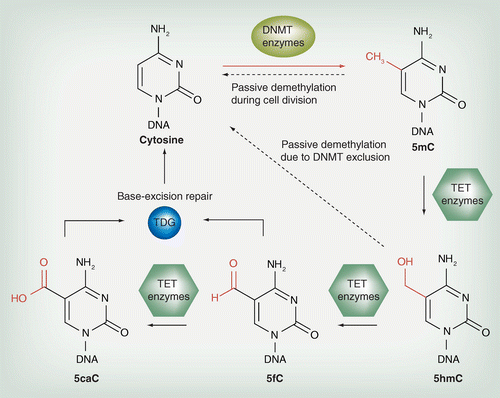
A, top) Bisulfite sequencing (BS-seq) cannot discriminate methylated C... | Download Scientific Diagram

Quantitative single cell 5hmC sequencing reveals non-canonical gene regulation by non-CG hydroxymethylation | bioRxiv

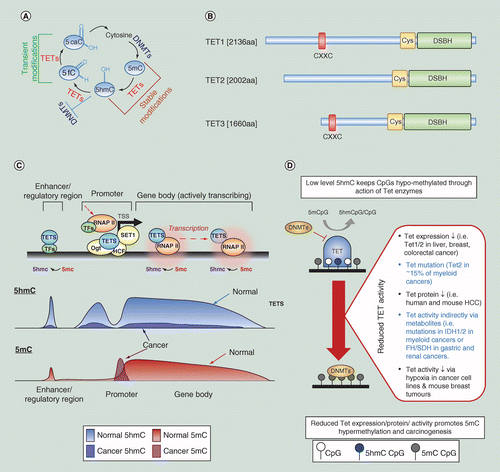
Epigenomic analysis of 5-hydroxymethylcytosine (5hmC) reveals novel DNA methylation markers for lung cancers - ScienceDirect

DeepH&M: Estimating single-CpG hydroxymethylation and methylation levels from enrichment and restriction enzyme sequencing methods | Science Advances

5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers | Cell Research

Jump-seq: Genome-Wide Capture and Amplification of 5-Hydroxymethylcytosine Sites | Journal of the American Chemical Society

A human tissue map of 5-hydroxymethylcytosines exhibits tissue specificity through gene and enhancer modulation | Nature Communications
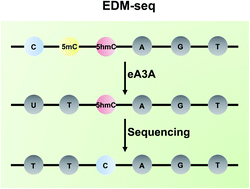
Bisulfite-free and single-nucleotide resolution sequencing of DNA epigenetic modification of 5-hydroxymethylcytosine using engineered deaminase - Chemical Science (RSC Publishing)

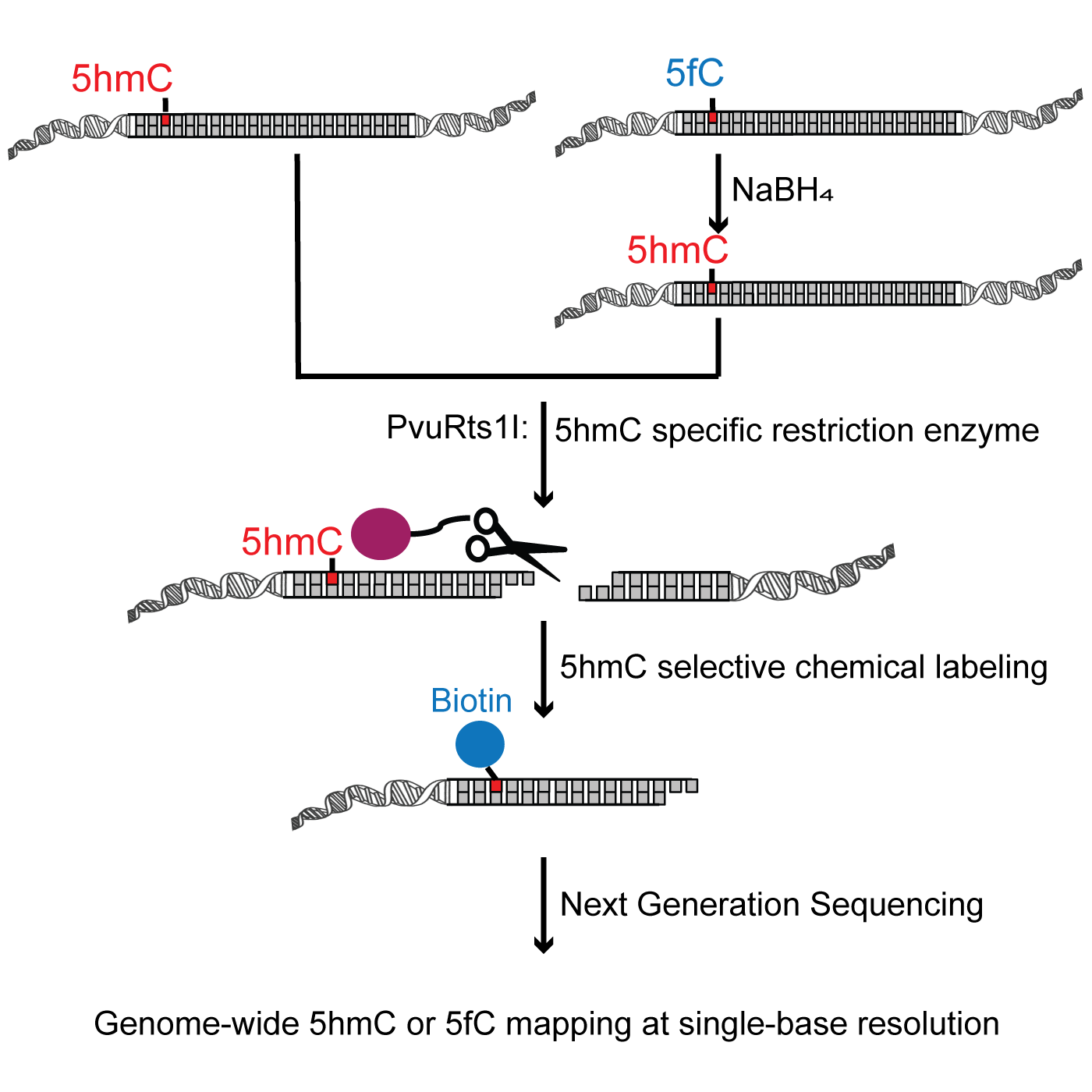
A Sensitive Approach to Map Genome-wide 5-Hydroxymethylcytosine and 5-Formylcytosine at Single-Base Resolution - ScienceDirect

Non-destructive enzymatic deamination enables single molecule long read sequencing for the determination of 5-methylcytosine and 5-hydroxymethylcytosine at single base resolution | bioRxiv