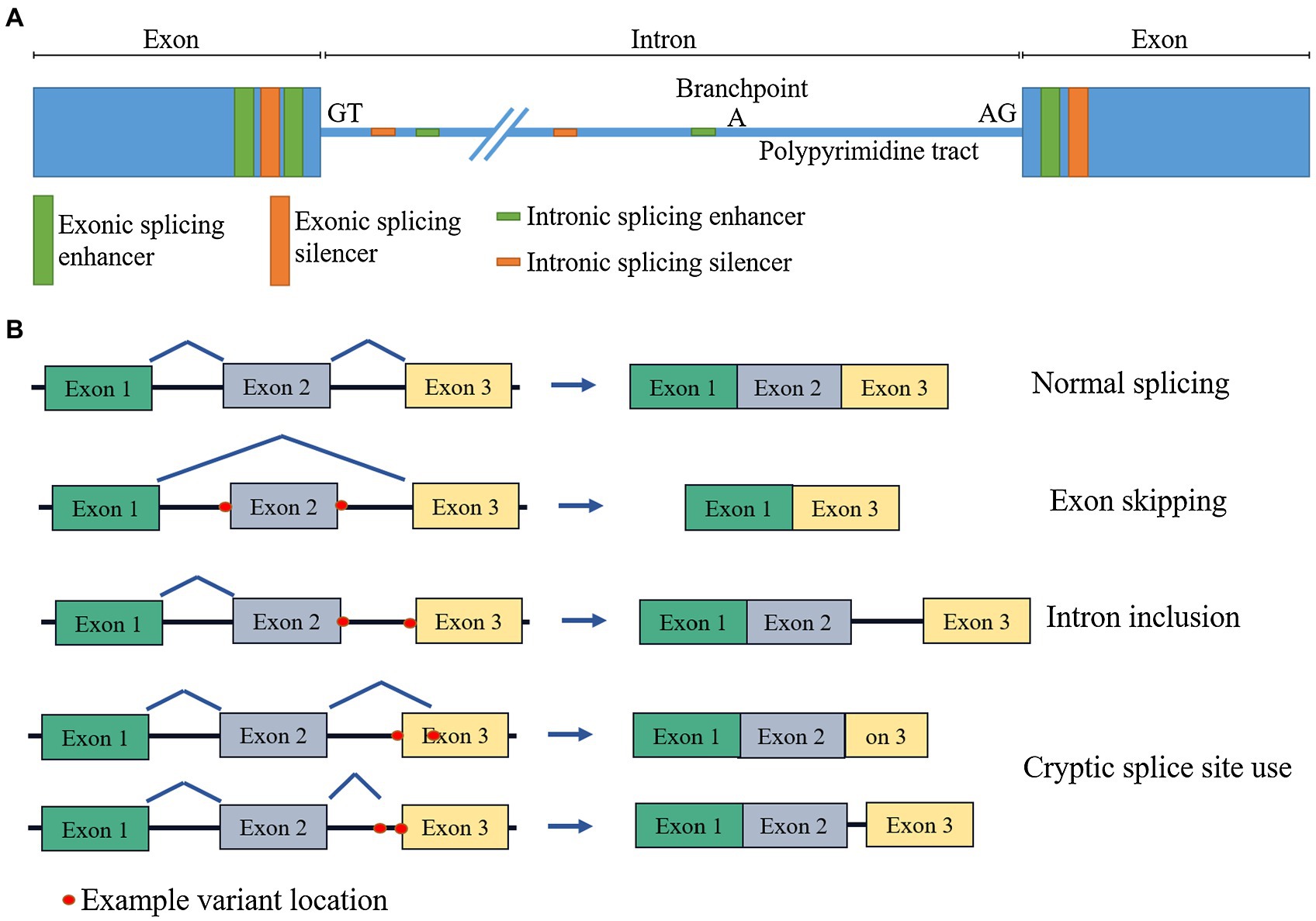
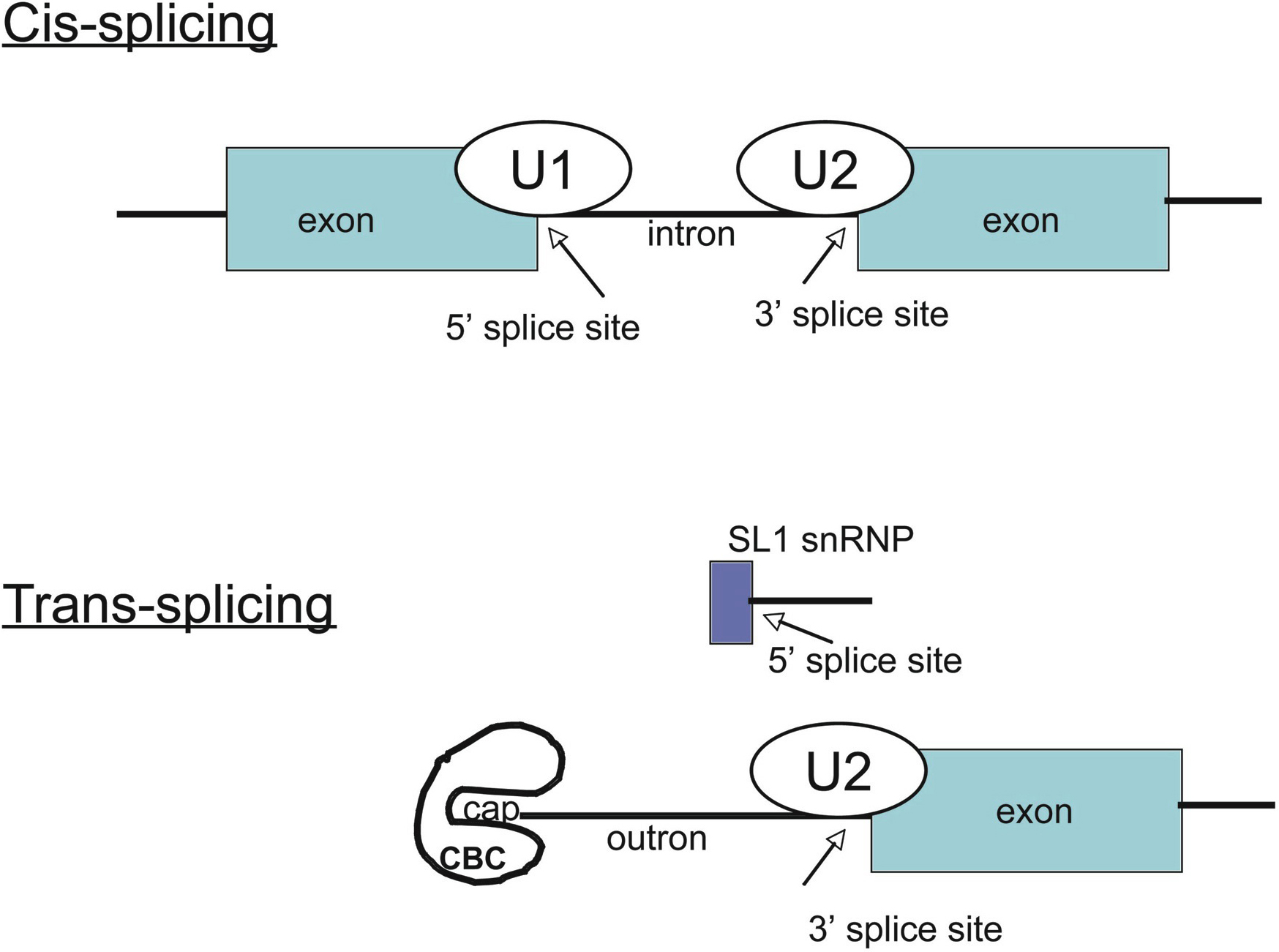
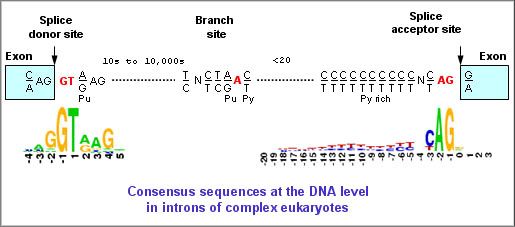
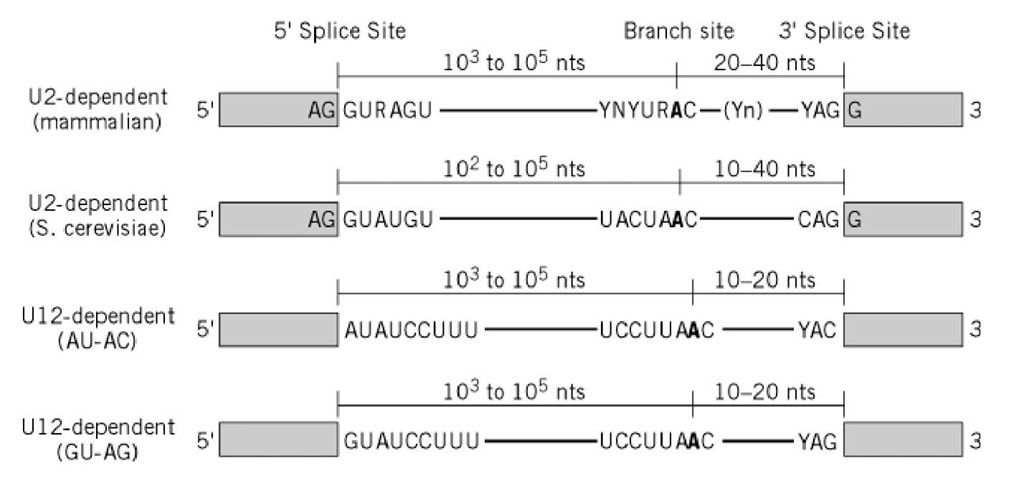
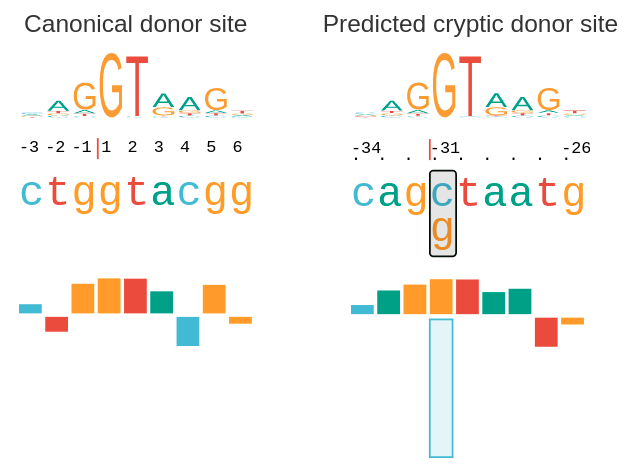
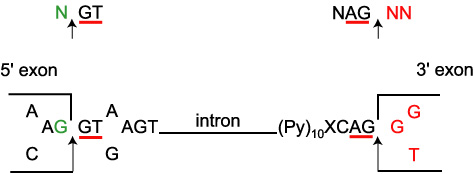
Before progressing from “exomes” to “genomes”… don't forget splicing variants | European Journal of Human Genetics

IJMS | Free Full-Text | Coupling and Coordination in Gene Expression Processes with Pre-mRNA Splicing
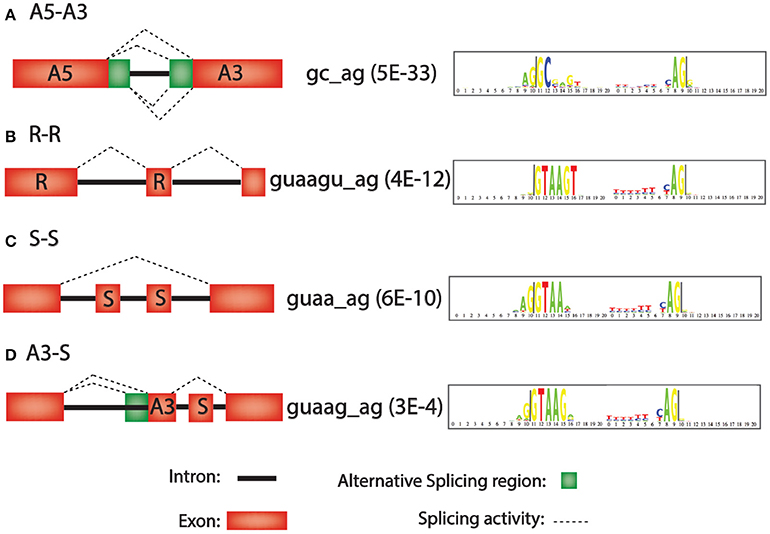
Frontiers | A Bioinformatics-Based Alternative mRNA Splicing Code that May Explain Some Disease Mutations Is Conserved in Animals
Oriented Scanning Is the Leading Mechanism Underlying 5′ Splice Site Selection in Mammals | PLOS Genetics

Cross talk between the upstream exon-intron junction and Prp2 facilitates splicing of non-consensus introns - ScienceDirect
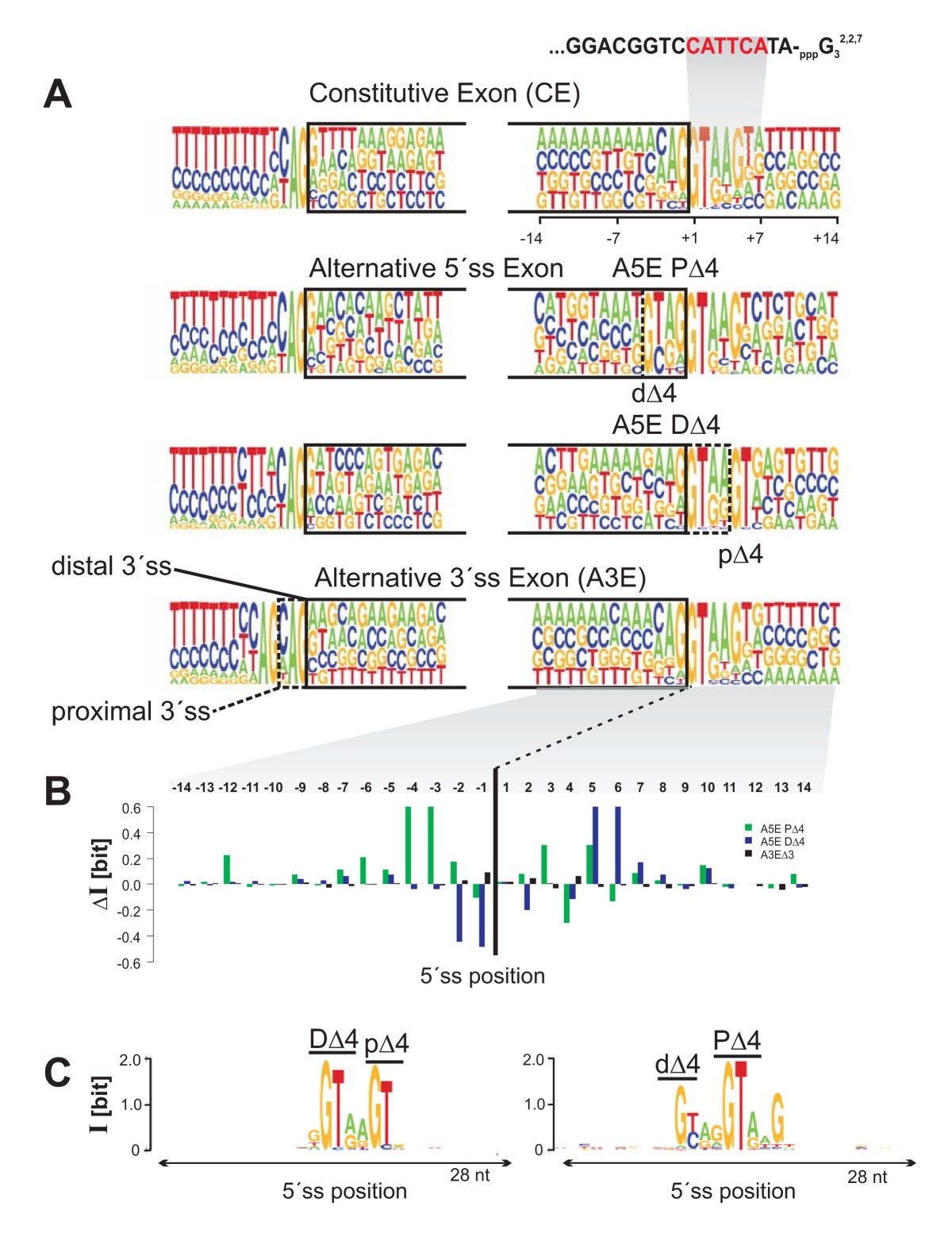
Comparative analysis of sequence features involved in the recognition of tandem splice sites | BMC Genomics | Full Text
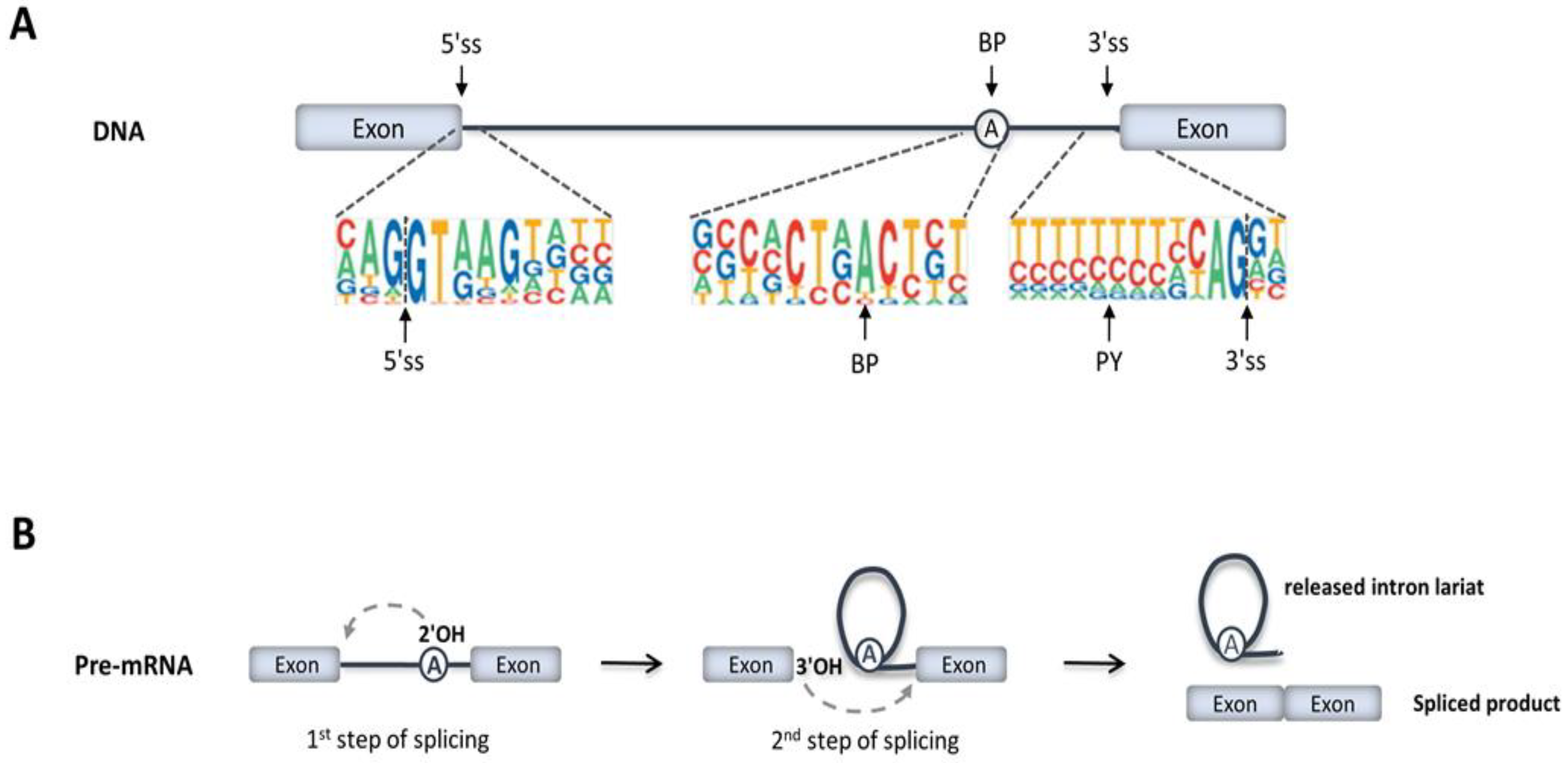
IJMS | Free Full-Text | RNA Splicing Defects in Hypertrophic Cardiomyopathy: Implications for Diagnosis and Therapy