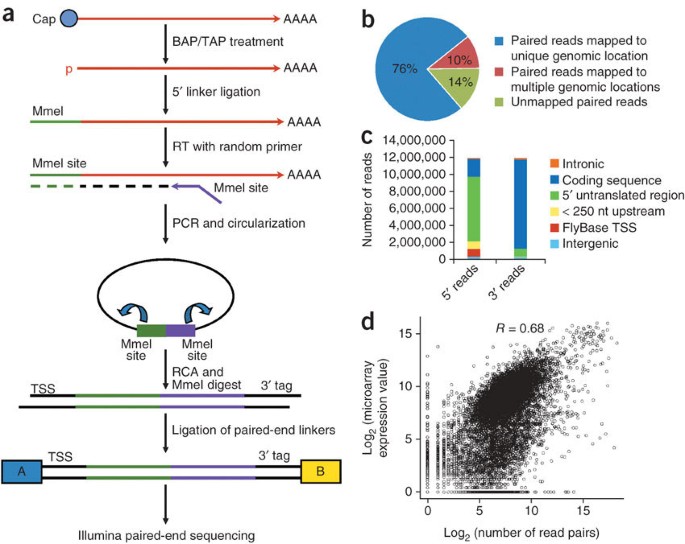
A paired-end sequencing strategy to map the complex landscape of transcription initiation | Nature Methods

Evaluation of Compatibility of 16S rRNA V3V4 and V4 Amplicon Libraries for Clinical Microbiome Profiling | bioRxiv

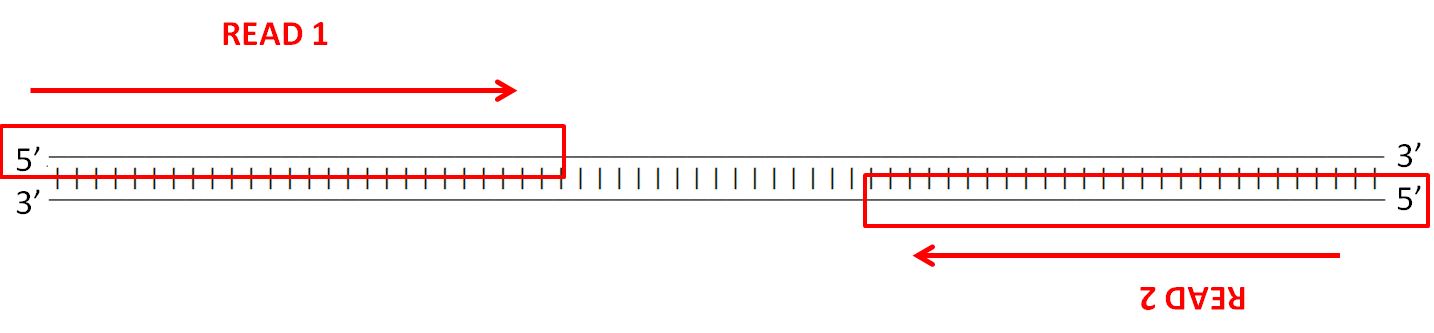
Paired-end sequencing (left) showing Read 1 and Read 2 primers starting... | Download Scientific Diagram

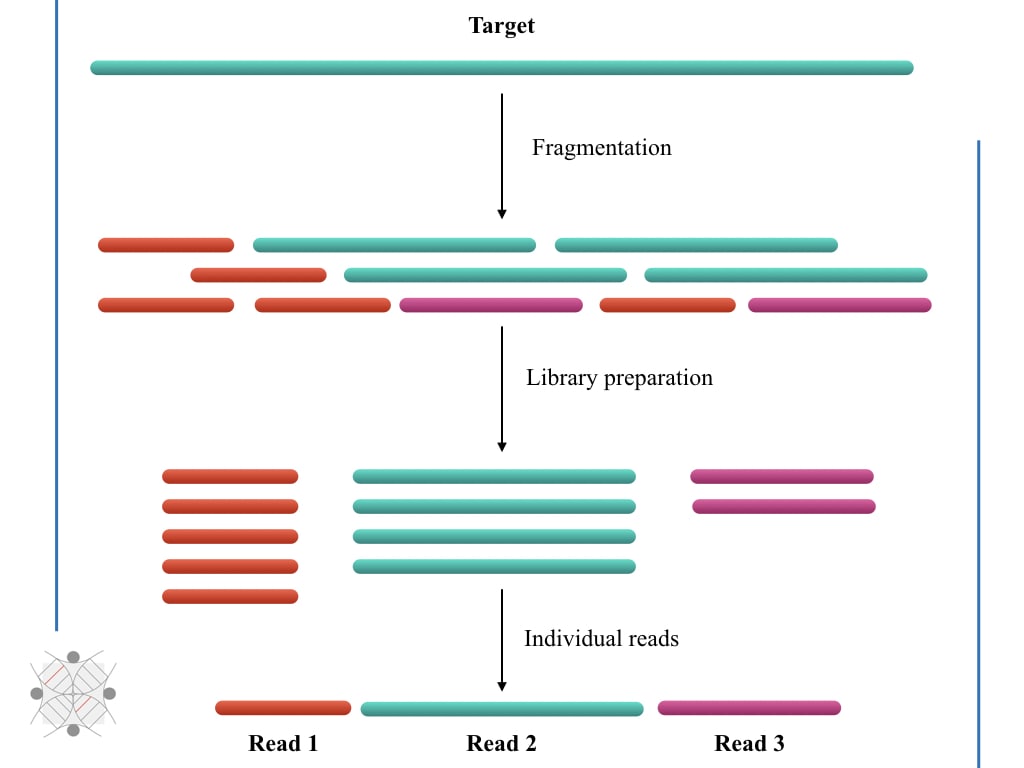
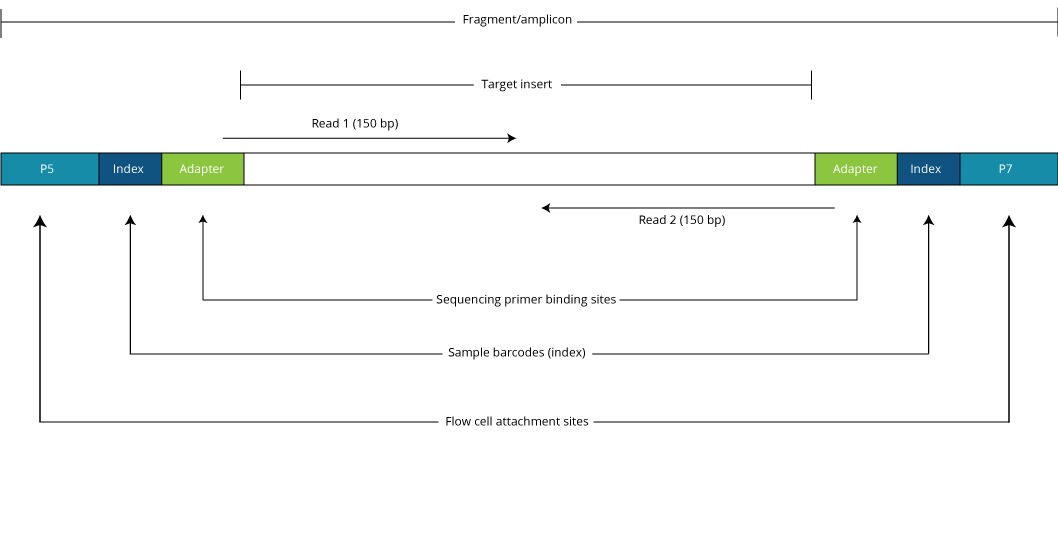
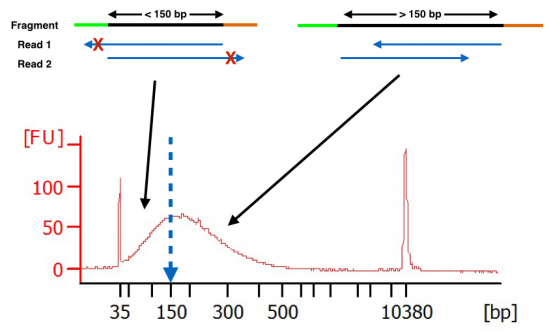
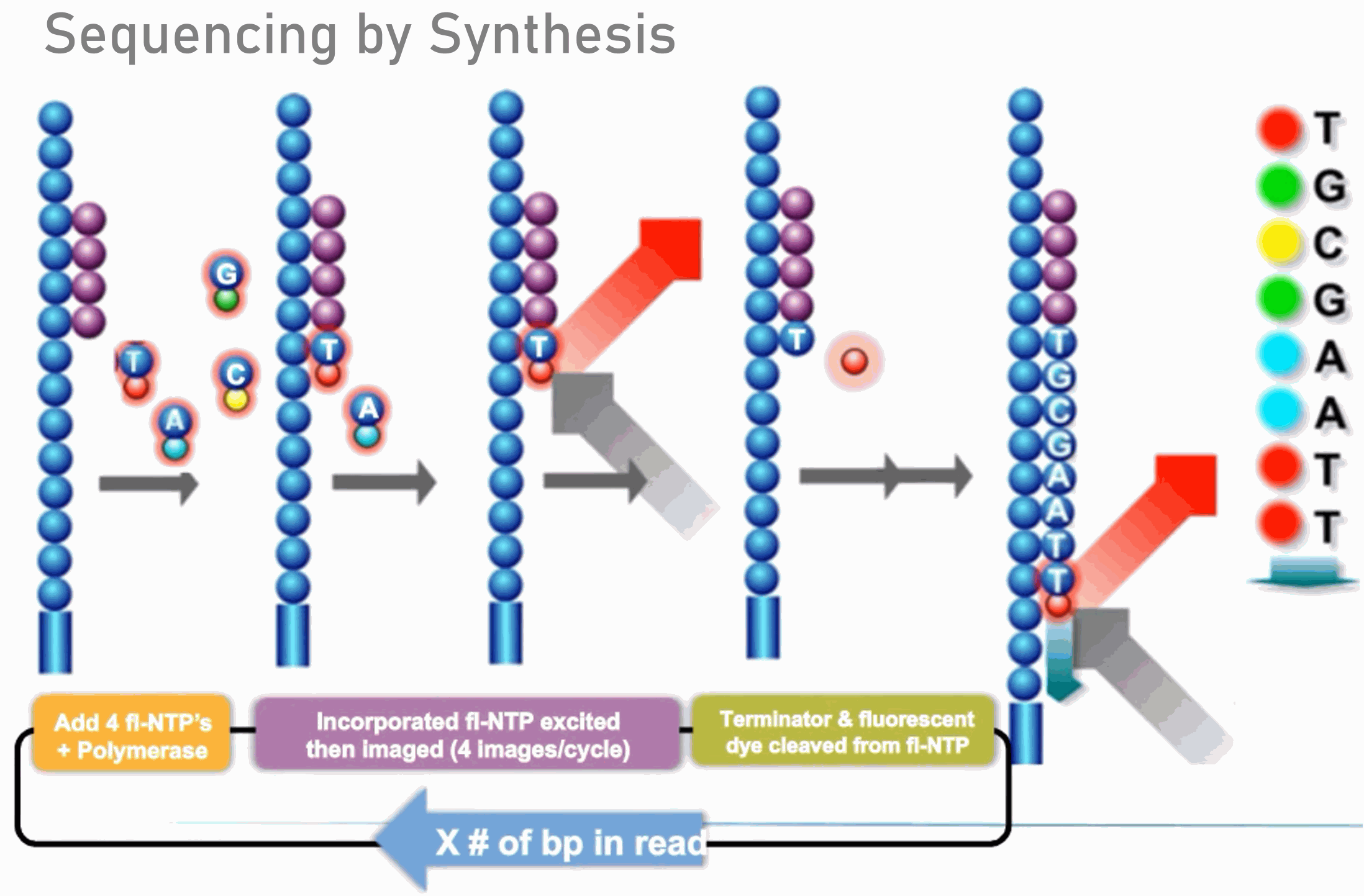
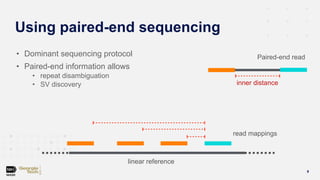
Compbio 020: Reads, fragments and inserts - what you need to know for understanding your sequencing data — Bad Grammar, Good Syntax
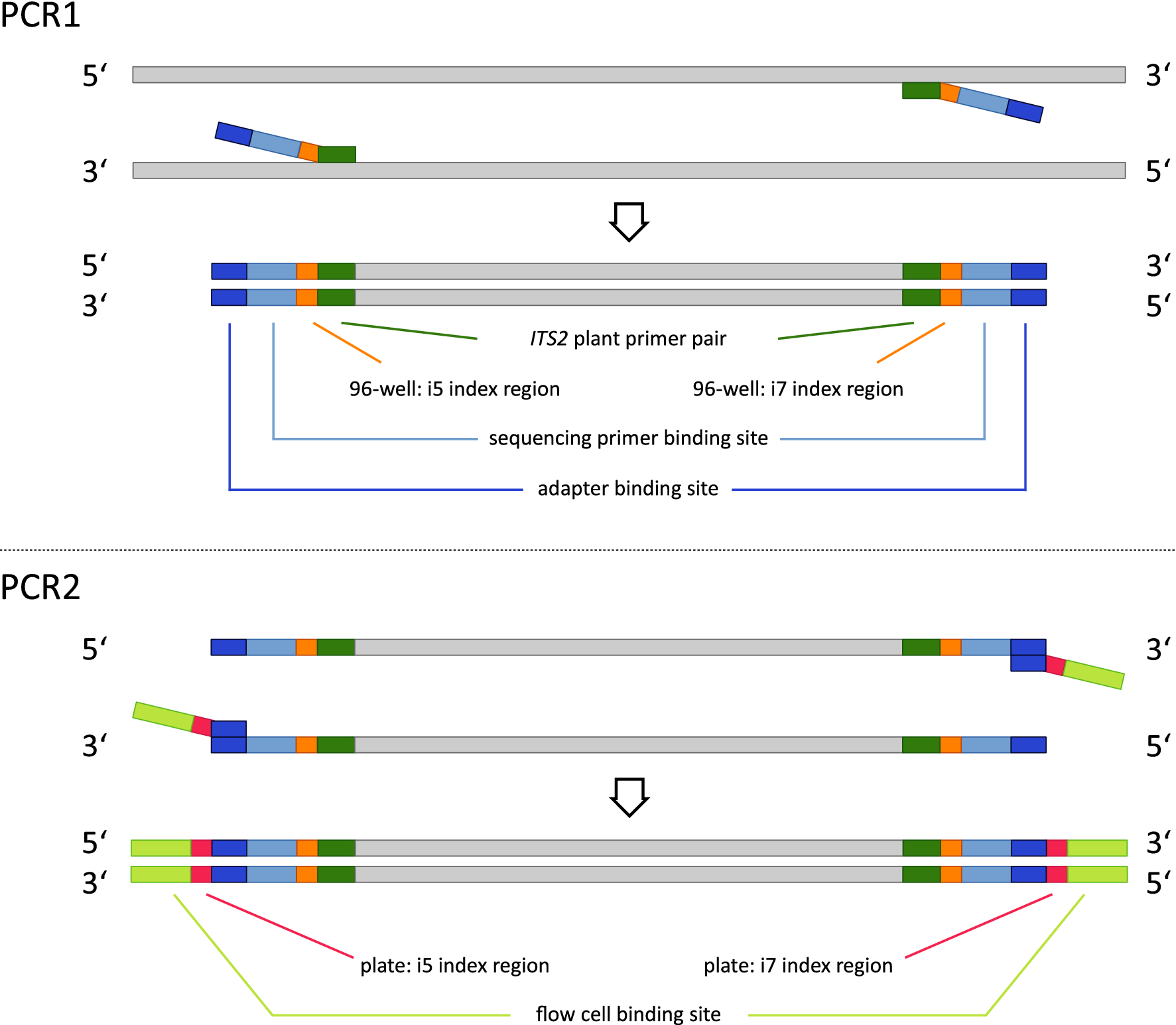
Handling of targeted amplicon sequencing data focusing on index hopping and demultiplexing using a nested metabarcoding approach in ecology | Scientific Reports
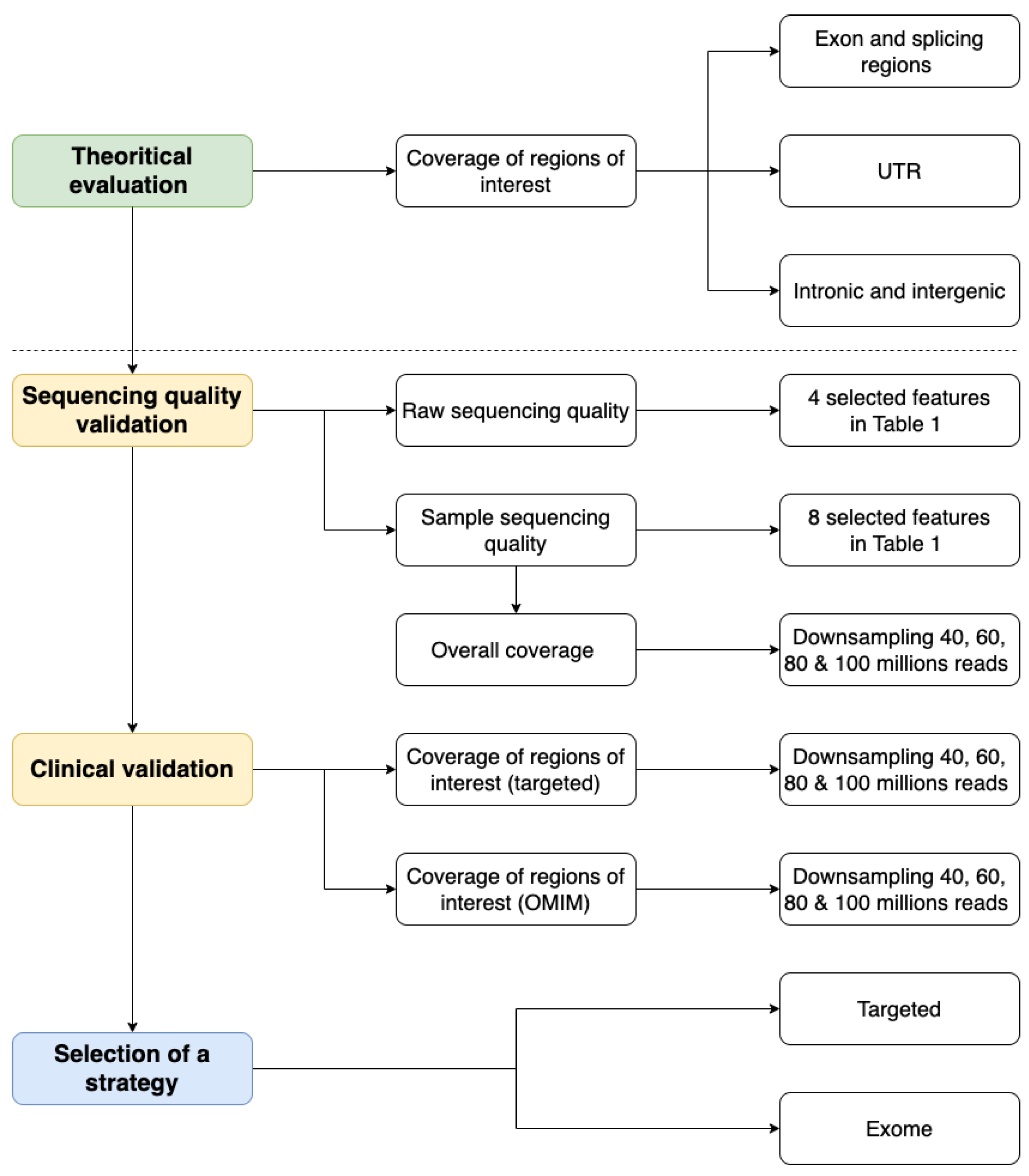
IJMS | Free Full-Text | Evaluating the Transition from Targeted to Exome Sequencing: A Guide for Clinical Laboratories