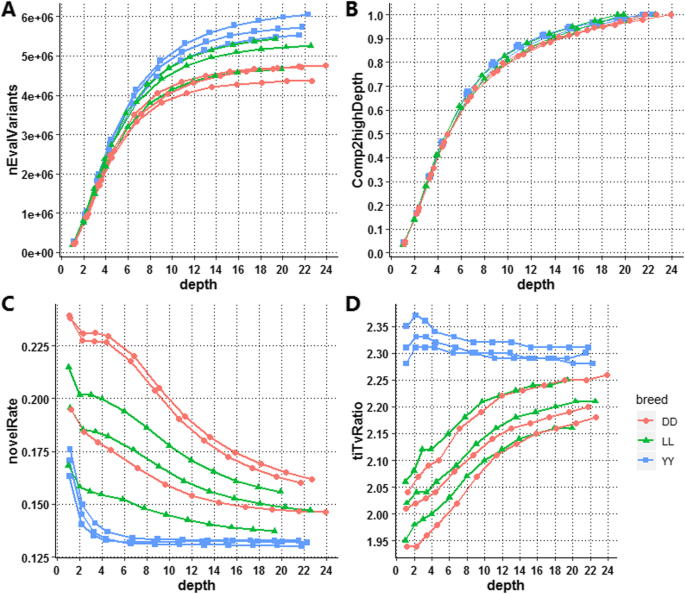
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text
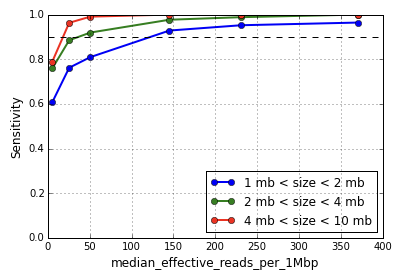
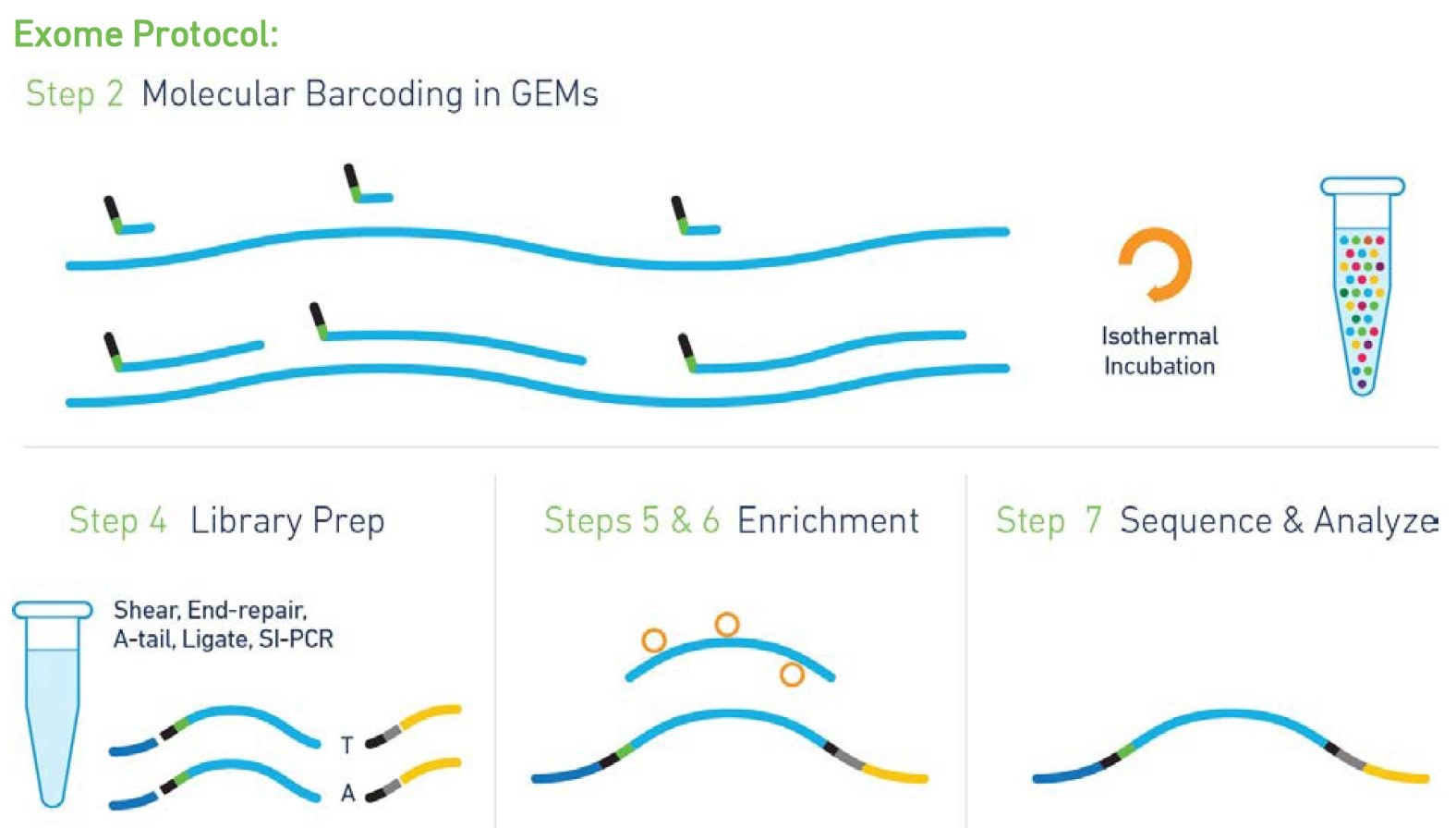
Impact of sequencing depth on copy number detection -Software -Single Cell CNV -Official 10x Genomics Support
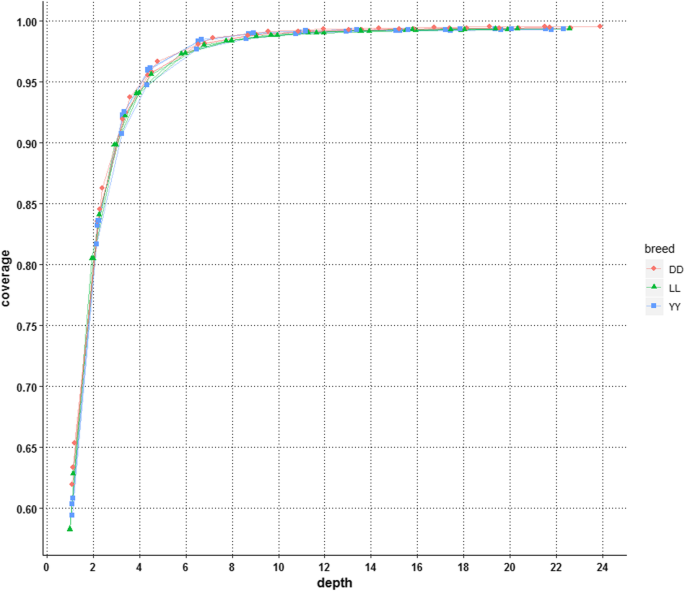
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text

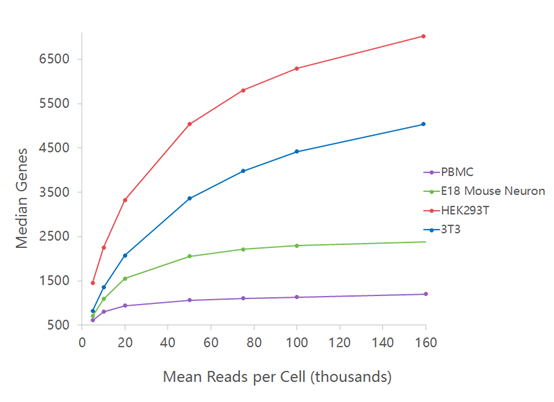
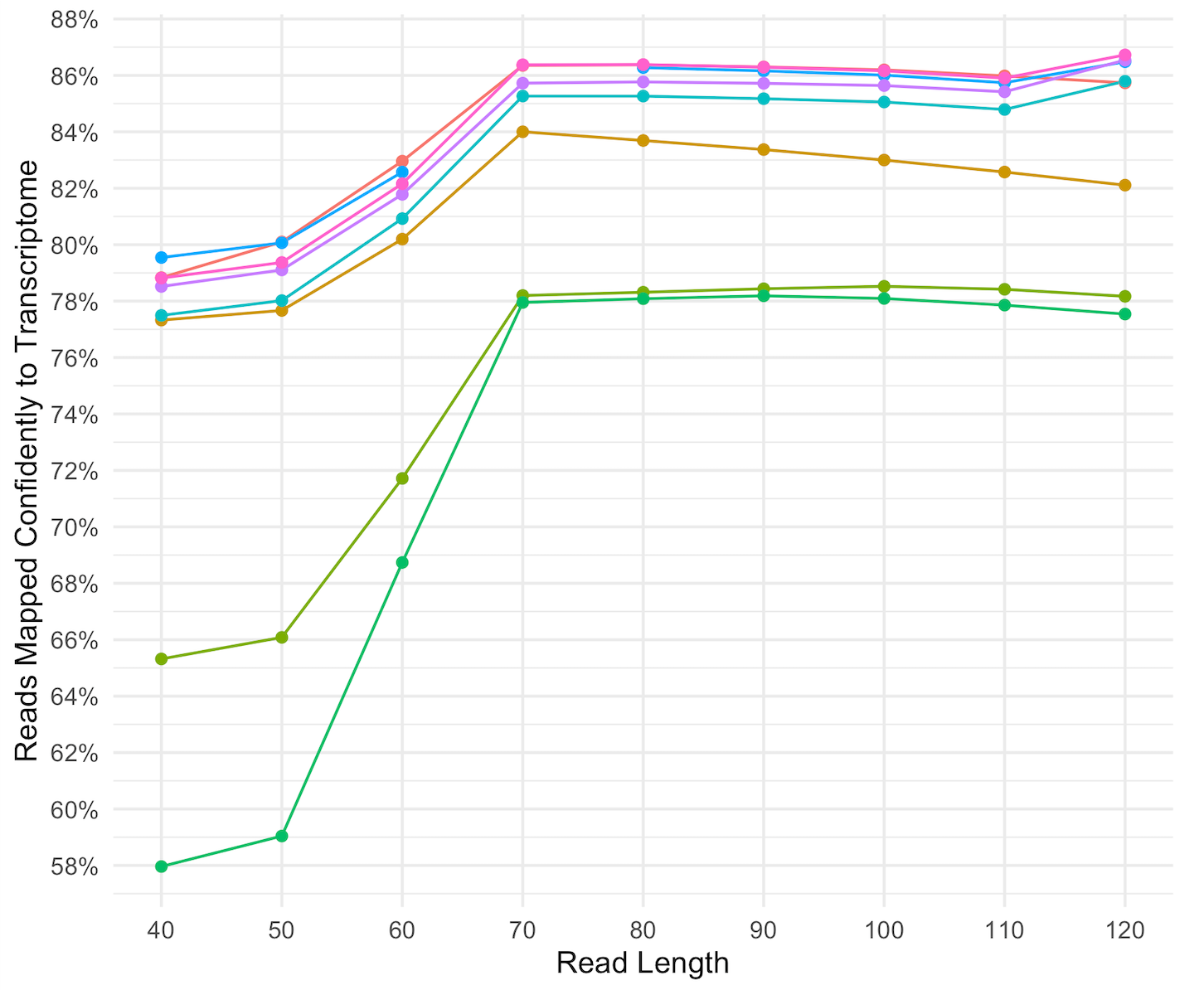
Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA- Seq Systems - ScienceDirect

Sample depth of coverage. Histogram of the mean sequencing read depth... | Download Scientific Diagram
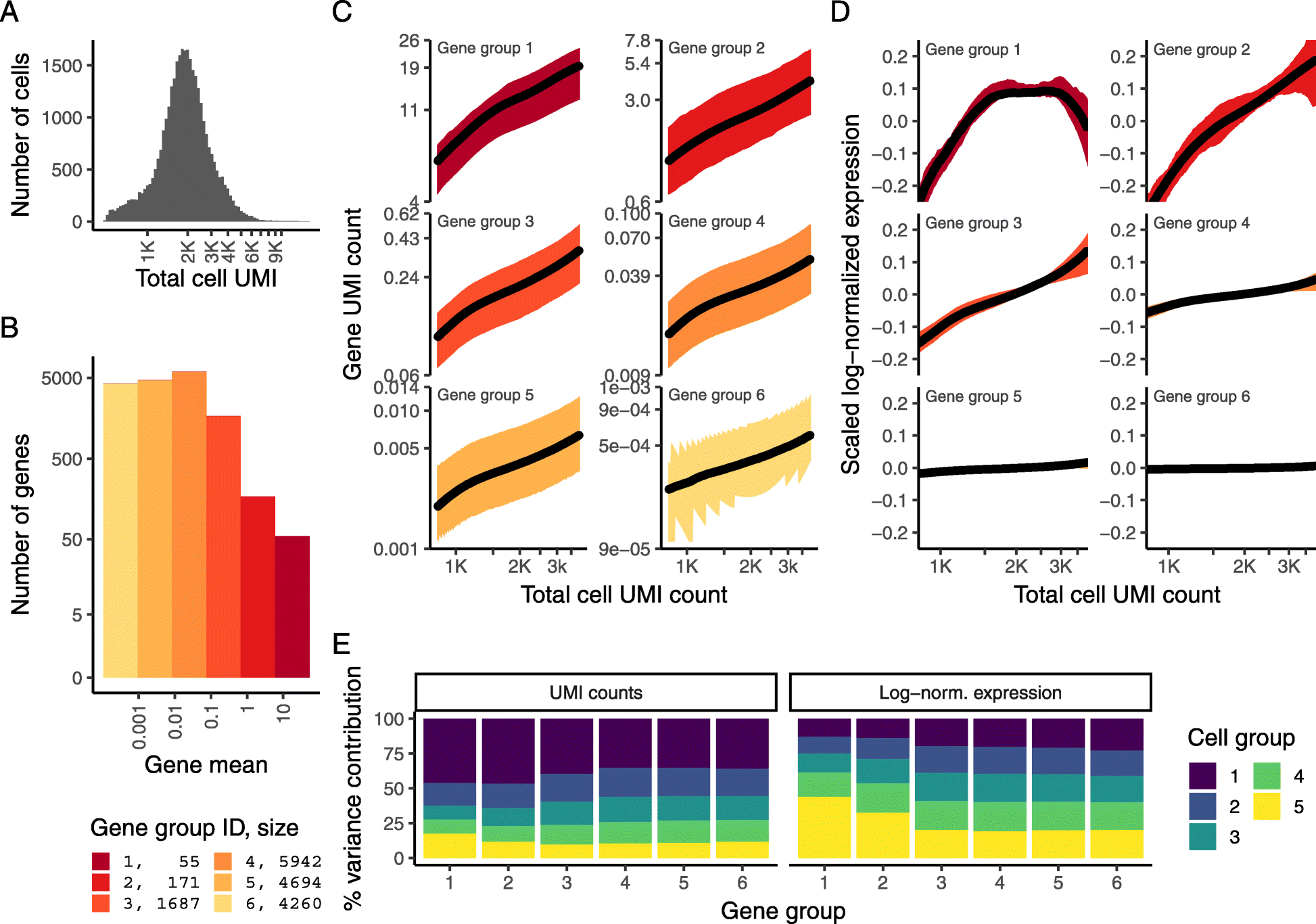
Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression | Genome Biology | Full Text